Mass spectrum data compression method
A technology of mass spectrometry data and compression method, which is applied in the direction of electronic digital data processing, special data processing application, digital data information retrieval, etc., can solve the problems of no significant increase in compression rate, poor software adaptability, and low compression rate, etc., to achieve Fast reading speed, small space, efficient data exchange and reading effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
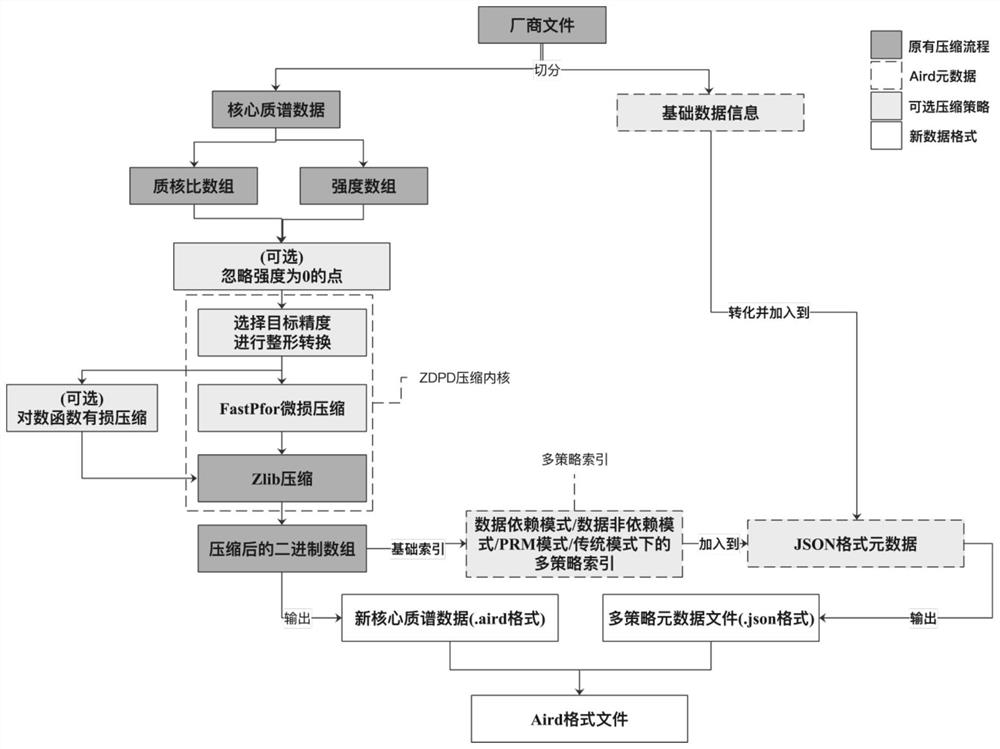
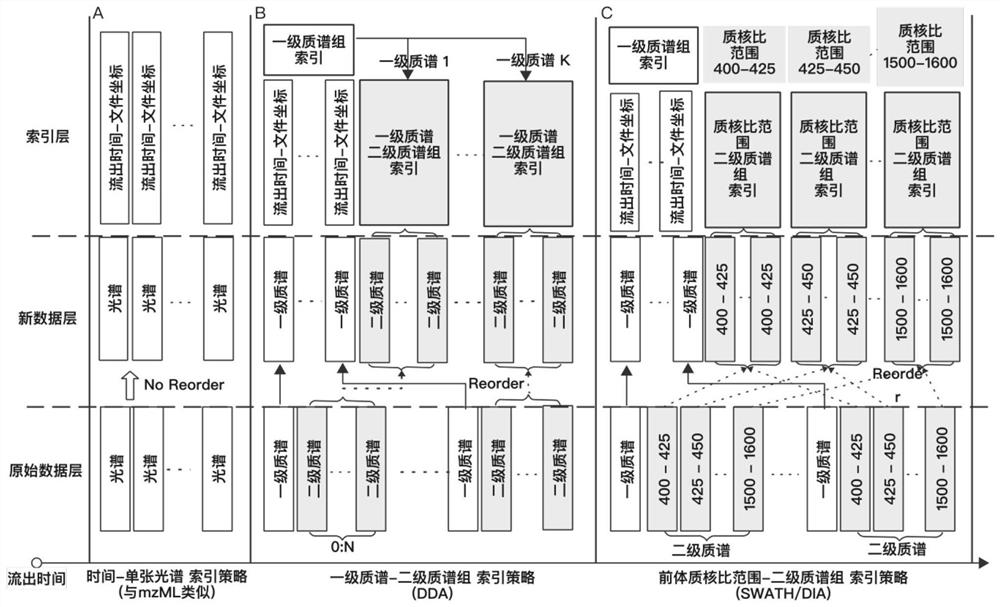
[0030] like figure 1 Shown, a kind of mass spectrometry data compression method, comprises the following steps:
[0031] S1. Segment the original mass spectrum data file, which is divided into mass spectrum data and basic metadata, wherein the mass spectrum data includes a mass-to-nucleus ratio array and an intensity array, and the mass-to-nucleus ratio array and the intensity array have the same length and are in one-to-one correspondence;
[0032] S2. The mass-to-core ratio array and the intensity array are compressed in the ZDPD compression kernel and converted into binary data; at the same time, the basic metadata information of the mass spectrum is saved in JSON format; after the end of step S1, it also includes deleting the point where the intensity information of the mass spectrum data is 0.
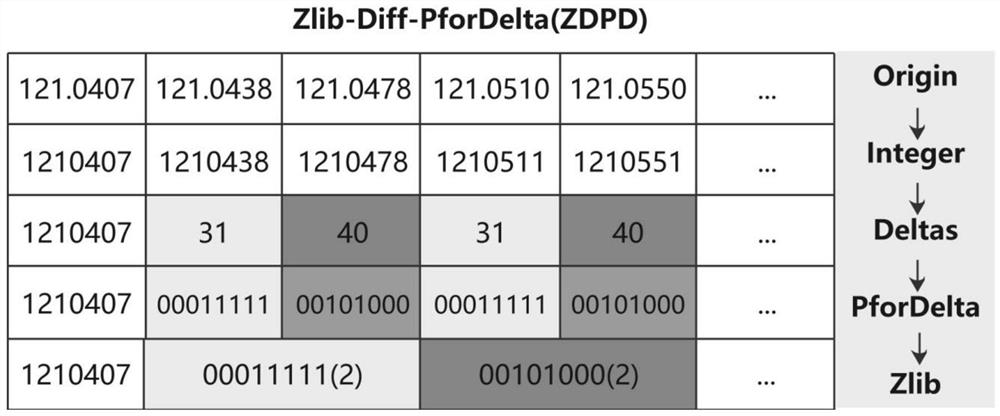
[0033] like figure 2 As shown, the algorithm principle and compression steps of ZDPD are as follows:
[0034] S21. Perform integer conversion according to the required target p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com