Primers, kit and detection method for identifying carcasses of silky chicken and black-bone chicken
A technology for black phoenix chickens and silky chickens, which is applied in the directions of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem that there is no effective method for identifying silk-feathered silky chickens and black phoenix chicken carcasses. and other issues, to achieve the effect of good social value, broad market application prospects and considerable economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
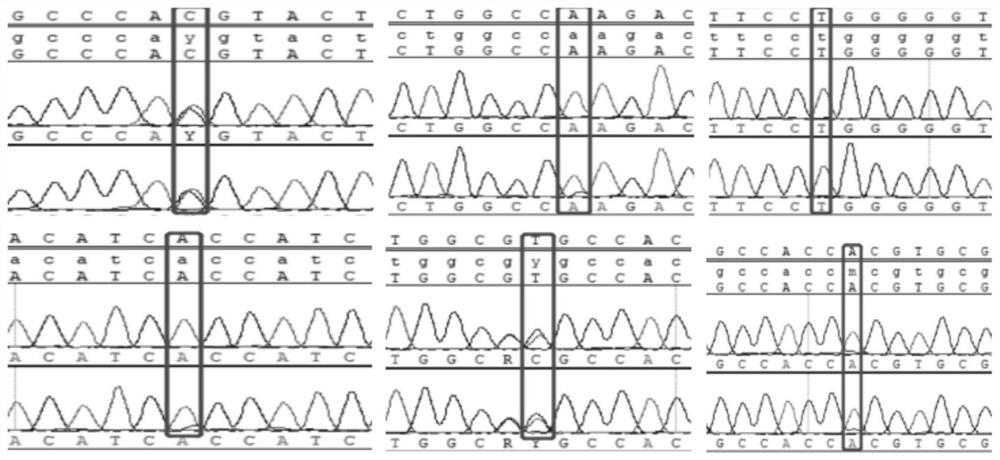
Image
Examples
Embodiment 1
[0028] Example 1 Identification of genome differences between silky silky chicken and black phoenix chicken
[0029] Step 1: Template Preparation
[0030] (1) Tissue collection: The skin samples of 24 silky chickens and 24 black-bone chickens were collected.
[0031] (2) DNA template preparation: about 50 mg of skin samples were collected, put into a centrifuge tube with 500 μL of liquid nitrogen, and the collected samples were ground with a tissue grinder. After the liquid nitrogen was evaporated to dryness, 200 microliters of TE was added, centrifuged at 1200 rpm / min, and the supernatant was extracted for later use.
[0032] Step 2: Type the target locus
[0033] (1) PCR amplification
[0034] PCR amplification primers are shown in Table 1:
[0035] Table 1 PCR amplification primers
[0036]
[0037] Note: "S1-CXF" is the sequencing primer corresponding to the PCR amplified fragment. Using additional primers for sequencing can save the step of PCR product purificati...
Embodiment 2
[0056] Reliability of Embodiment 2 Single-blind Detection and Judgment Method
[0057] In this example, a single-blind method is adopted to determine the source of the species of the samples of known species according to the molecular detection results.
[0058] Step 1: Template Preparation
[0059] (1) Tissue collection: The skin tissues of 12 silky chickens and 12 black-bone chickens were collected.
[0060] (2) DNA template preparation: about 50 mg of skin samples were collected, put into a centrifuge tube with 500 μL of liquid nitrogen, and the collected samples were ground with a tissue grinder. After the liquid nitrogen was evaporated to dryness, 200 microliters of TE was added, centrifuged at 1200 rpm / min, and the supernatant was extracted for later use.
[0061] Step 2: Type the target locus
[0062] (1) PCR amplification
[0063] PCR amplification primers are shown in Table 7:
[0064] Table 7 PCR amplification primers
[0065]
[0066] Note: "S1-CXF" is the se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com