Kit for detecting phenotypes of circulating tumor cells
A tumor cell and kit technology, applied in the field of immunoassay detection, can solve the problems of missed CTC screening, inaccurate results, false positives, etc., and achieve the effects of improved sensitivity, high reaction efficiency, and sensitive detection.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
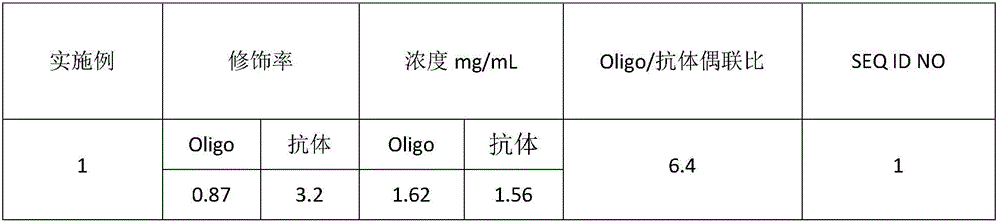
Embodiment 1
[0040] Embodiment 1E-cadherin-oligonucleotide probe
[0041] (1) Aldehyde modification of oligonucleotide probes
[0042] 50 nmol of Oligo1 (oligonucleotide) was prepared into a solution with 0.1 M pH 7.4 phosphate buffer. Weigh 500nmol of SFB (N-succinimidyl 4-formylbenzoate), dissolve it with anhydrous DMF (N,N-dimethylformamide), react at room temperature for 2.5h, and purify through a column to obtain Oligo -FB (aldehyde modified oligonucleotide).
[0043] Detection of the concentration of Oligo-FB: detection of A by Nanodrop spectrophotometer 260 value, the calculated concentration of Oligo-FB was 0.65nmol / μL.
[0044] Detection of aldehyde group modification rate: Use quantitative 2-hydrazinopyridine-2-hydrochloride solution to detect the aldehyde group modification rate. Take the above-mentioned Oligo-FB and add it to the 2-hydrazinopyridine-2-hydrochloride solution, shake and mix well, and react at 37°C for 1 hour. The absorbance value at 360nm detected by Nanodrop...
Embodiment 2
[0057] The QPCR amplification specific detection of embodiment 2Oligo molecule
[0058] Oligo1-13 molecules were diluted to three concentrations of 25000 molecules / μl, 83333 molecules / μl and 250000 molecules / μl. Then, taking Oligo1-3 as an example, mix Oligo1-3 at the same concentration to obtain mixed samples A, B, and C, as shown in Table 2.
[0059] Table 2 Example 2 sample formula
[0060]
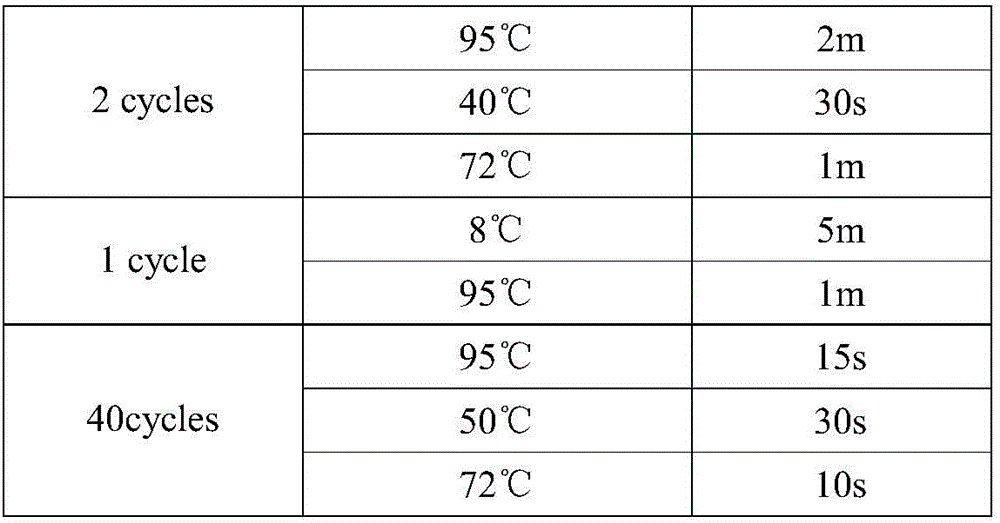
[0061] The above samples were configured with QPCR amplification solution according to Table 3 to obtain a QPCR amplification kit, and then Oligo1-3 was subjected to QPCR amplification according to the procedures shown in Table 4, and Oligo1-3 in mixed samples A, B, and C Each was subjected to QPCR amplification, using the extended Oligo1-3 as a template, and the Ct values of the amplification results are shown in Table 5. Among them, the extension primer is RT-P, its 3' end is complementary to the 3' end of the oligonucleotide, the 5' end can form a hairpin structure, and the m...
Embodiment 3
[0076] Embodiment 3 makes standard curve
[0077] The same result can be obtained with Oligo1-13. In order to simplify the process, the following examples use Oligo1-3 as an example.
[0078] According to the steps of Example 1, Oligo2 was modified with 5 times the molar equivalent of SFB to obtain Oligo-FB, and EpCAM was hydrazine-modified with 10 times the molar equivalent of SANH to obtain EpCAM-SANH, and the molar ratio was 10:1 EpCAM-Oligo2 was obtained after reacting Oligo-FB with EpCAM-SANH at room temperature for 16 hours.
[0079] According to the steps of Example 1, Oligo3 was modified with 20 times the molar equivalent of SFB to obtain Oligo-FB, and Cadherin-11 was hydrazino-modified with 50 times the molar equivalent of SANH to obtain Cadherin-11-SANH. The molar ratio Cadherin-11-Oligo3 was obtained after 8:1 Oligo-FB reacted with Cadherin-11-SANH at room temperature for 24 hours.
[0080] E-cadherin-Oligo1, EpCAM-Oligo2 and Cadherin-11-Oligo3 were subjected to Q...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com