D-lactic acid dehydrogenase of Sporolactobacillus inulinus, coding gene and application thereof
A lactate dehydrogenase and encoding technology, applied in the field of D-lactate dehydrogenase, can solve problems such as inability to obtain a single configuration compound, impure product, complex technology, etc., and achieve the effect of broad industrial application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
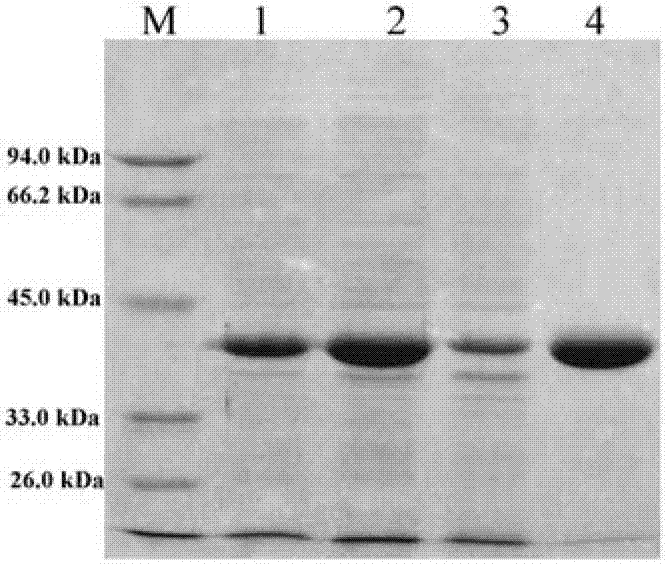
[0052] Embodiment 1, the acquisition of D-lactate dehydrogenase gene and its protein
[0053] 1. Cloning of D-lactate dehydrogenase gene
[0054] After sequencing the entire genome of Lactobacillus inulinus (Sporolactobacillus inulinus) CASD CGMCC No.2185 and annotating the genome information, a gene that may encode lactate dehydrogenase was obtained, and the following primers P1 and P2 were designed according to the gene sequence.
[0055] P1: 5'-GG TCATGA AAATCATTATGTTCAG-3' (the underline is the recognition sequence of the restriction site BspH I, the 5th-24th is the 1st-20th of the sequence 2)
[0056] P2: 5'-GC AAGCTT GTTTTCTACAGCTACTTTGTT-3' (underlined is the recognition sequence of the restriction site Hind III, and the following sequence is the reverse complementary sequence of the 985-1005th position of sequence 2)
[0057] Using the total gene of Sporolactobacillus inulinus (Sporolactobacillus inulinus) CASD CGMCC No.2185 as a template, and using P1 and P2 as p...
Embodiment 2
[0069] The enzymatic characteristic identification of embodiment 2, embodiment 1 gained object protein
[0070] Identify the enzymatic properties of the resulting protein of interest in Example 1 with the following reactions:
[0071] (Phenyl)pyruvate+NAD(P)H+H + →D-(phenyl)lactic acid+NAD(P) +
[0072] With 20mM pyruvate or 20mM phenylpyruvate as substrate, 0.2mM NADH or 0.2mM NADPH as coenzyme, and 15μg D-lactate dehydrogenase were added to 100mM phosphate buffer (pH5.5) and mixed (the concentrations of the above substances are the final concentrations in 100mM phosphate buffer), and react at 30°C for 1 hour. After the reaction is finished, the contents of D-lactic acid and L-lactic acid, or D-phenyllactic acid and L-phenyllactic acid in the reaction liquid are detected.
[0073] The results show that: the target protein obtained in Example 1 can use NADH as a coenzyme, and can also use NADPH as a coenzyme, specifically:
[0074] Using NADH as a coenzyme to catalyze pyr...
Embodiment 3
[0087] Embodiment 3, utilize D-lactate dehydrogenase to produce D-phenyllactate
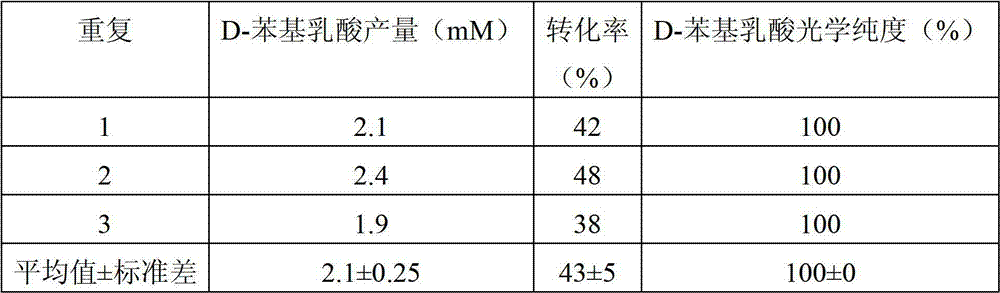
[0088] Add 15 μg of the target protein (D-lactate dehydrogenase) obtained in Example 1, 5 mM phenylpyruvate (substrate), and 2.1 mM NADH (coenzyme) to 100 mM phosphate buffer (pH5.5 ) (the concentrations of the above substances are the final concentrations in 100mM phosphate buffer), and react at 30°C for 1 hour. After the reaction, the content of D-phenyllactic acid and L-phenyllactic acid in the reaction solution was detected. Calculate substrate conversion and optical purity of D-phenyllactate. The experiment was repeated three times, and the results were averaged.
[0089] The results are shown in Table 2. D-phenyllactic acid at a concentration of 2.1 mM was detected in the reaction solution, but the presence of L-phenyllactic acid was not detected.
[0090] Table 2 Results of 3 repeated experiments using D-lactate dehydrogenase cells to produce D-phenyllactate (1)
[0091]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com