Method for identifying Frankliniella occidentalis species by utilizing PCR (Polymerase Chain Reaction)-RFLP (Restriction Fragment Length Polymorphism) technology
A technology of PCR-RFLP and western flower thrips, applied in the field of agricultural biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
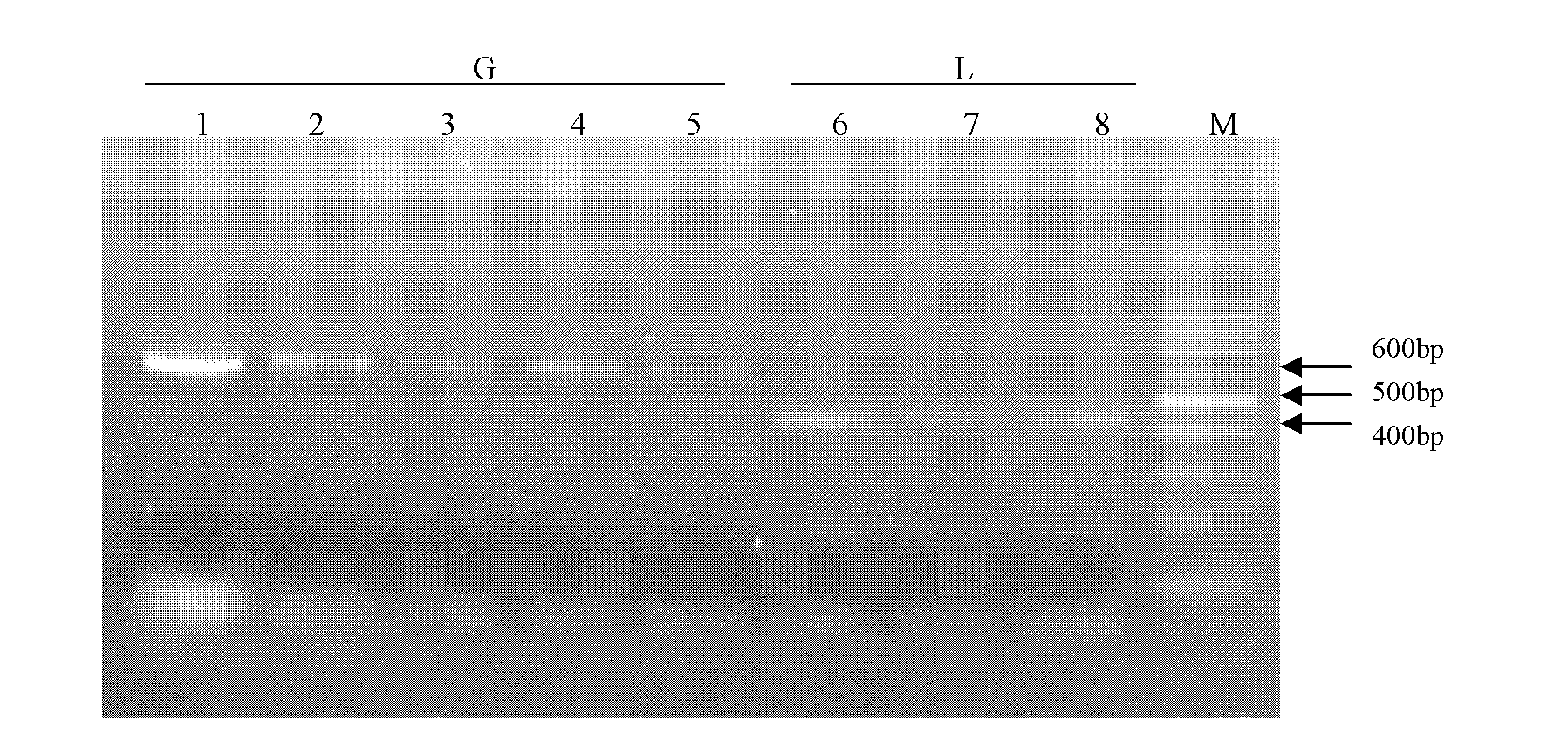
[0033] Analysis of the sample material of Thrips occidentalis
[0034] (1) Extraction of Western Flower Thrips Genomic DNA
[0035] Place a single thrips individual in a 0.2ml centrifuge tube containing 60 μl of alkaline lysate: 50 mmol L -1 Tris-HCl (pH8.0), 20mmol·L -1 NaCl, 1mmol L -1 EDTA and 1% SDS were thoroughly ground and homogenized with a sealed pipette tip, placed in a water bath at 65°C for 15 minutes, and then at 95°C for 10 minutes to prepare a genomic DNA solution of Thrips occidentalis.
[0036] (2) PCR amplification of the COI gene of Thrips occidentalis
[0037] The COI gene was amplified by PCR using the genomic DNA of Thrips occidentalis as a template to obtain the PCR product;
[0038] The PCR amplification system is:
[0039] Genomic DNA: 3μl; 20μM primer: 0.5μl; 5U / μl Taq enzyme: 0.5μl; 10×Taq Buffer: 5μl;
[0040] 10mM dNTP: 1 μl; ddH 2 O supplemented to 50 μl;
[0041] Amplification primer sequences are as follows:
[0042]Sense primer: 5'GGAT...
Embodiment 2
[0048] A kind of method utilizing PCR-RFLP technology to distinguish western flower thrips strain, the steps are as follows:
[0049] (1) Extraction of Western Flower Thrips Genomic DNA
[0050] Place a single thrips individual in a 0.2ml centrifuge tube containing 60 μl of alkaline lysate: 50 mmol L -1 Tris-HCl (pH8.0), 20mmol·L -1 NaCl, 1mmol L -1 EDTA and 1% SDS were thoroughly ground and homogenized with a sealed pipette tip, placed in a water bath at 65°C for 15 minutes, and then at 95°C for 10 minutes to prepare a genomic DNA solution of Thrips occidentalis.
[0051] (2) PCR amplification of the COI gene of Thrips occidentalis
[0052] The COI gene was amplified by PCR using the genomic DNA of Thrips occidentalis as a template to obtain the PCR product;
[0053] The PCR amplification system is:
[0054] Genomic DNA 2μl, 20μM primer 0.4μl, 5U / μl Taq enzyme 0.2μl, 10×Taq Buffer 2μl,
[0055] 0.4 μl of 10 mM dNTPs, ddH 2 O supplemented to 20 μl;
[0056] Amplificati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com