Method for establishing colon cancer prognosis prediction model
A technique for predicting models and establishing methods, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Step 1: Detection of SPARCL1 and P53 proteins in colorectal cancer tissues
[0021] Immunohistochemical methods were used to detect the expression of P53 and SPARCL1 in the surgically resected colorectal cancer tissues of patients.
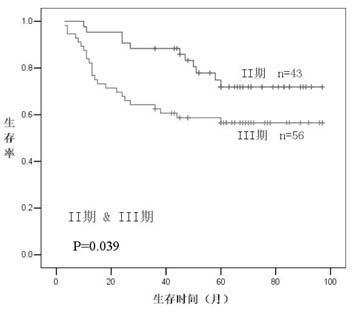
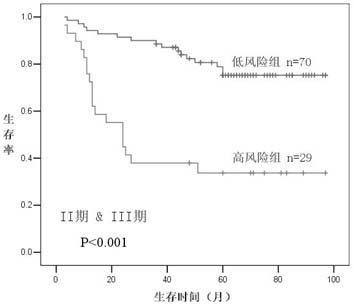
[0022] The 131 colorectal cancer tissue wax blocks were selected from surgically resected tissues of colorectal cancer patients admitted to the Second Affiliated Hospital of Zhejiang University School of Medicine from 1999 to 2004 (23 cases of stage I, Period 43 cases, Period 56 cases, 9 cases in the first stage), and the diagnosis was confirmed by postoperative pathology. The clinicopathological data of all patients were registered in the form of follow-up form after operation, and their survival, recurrence and metastasis were followed up for more than 36 months by letter or telephone every year. Check again before the experiment started.
[0023] All tissue wax block specimens were from the tissue wax blocks archived in the Patholo...
Embodiment 2
[0033] 1. The role of SPARCL1 / P53 model in judging the prognosis of colorectal cancer patients (all stages)
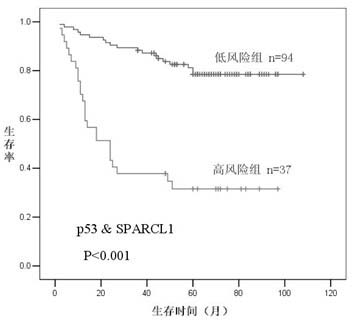
[0034] figure 1 Survival curves of patients (all stages) with different discriminant results of the SPARCL1 / P53 prognostic model are shown. The Kaplan-Meier method validation showed that the SPARCL1 / P53 prediction model could divide 131 patients into two groups with great differences in survival time (P<0.001), namely good prognosis (predicted value = 0) and bad prognosis (predicted value = 0). 1), the estimated median survival time of the patients in the good prognosis group was 91.676 months, and the estimated median survival time of the patients in the bad prognosis group was 41.928 months.
[0035] . Predictive effect of SPARCL1 / P53 model on postoperative recurrence and metastasis of colorectal cancer patients
[0036] Among these patients, the incidence of postoperative recurrence and metastasis in the good prognosis group (predicted value = 0) was 22.34%, whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com