Genes coding for tomato beta-galactosidase polypeptides
a technology of beta-galactosidase and polypeptides, which is applied in the field of genes coding for tomato beta-galactosidase polypeptides, can solve the problem of elusive cloning of the corresponding gen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNA Extraction
[0081] Tomato (Lycopersicon esculentum Mill., cv. ‘rutgers’) plants were grown in a greenhouse using standard cultural practices. The ripening mutants, ripening inhibitor (rin), non-ripening (nor) and never ripe (Nr) (Tigchelaar et al. 1978. Hort. Science 13: 508-513), were all in the ‘Rutgers’ background. Flowers were tagged at anthesis and fruit were harvested according to the number of days post-anthesis (dpa) or based on their surface color using ripeness stages as previously described (Mitcham et al. 1989. Plant Physiol. 89: 477-481), the complete disclosure of which is hereby fully incorporated herein by reference. For gene expression studies, a variety of leaf, flower, and stem tissues were harvested from greenhouse-grown plants and roots were harvested from seedlings grown in basal tissue culture medium for 4 weeks after seed germination.
[0082] Fruits were processed immediately after harvest in the greenhouse by chilling on ice, excising the various tissues a...
example 2
RT-PCR
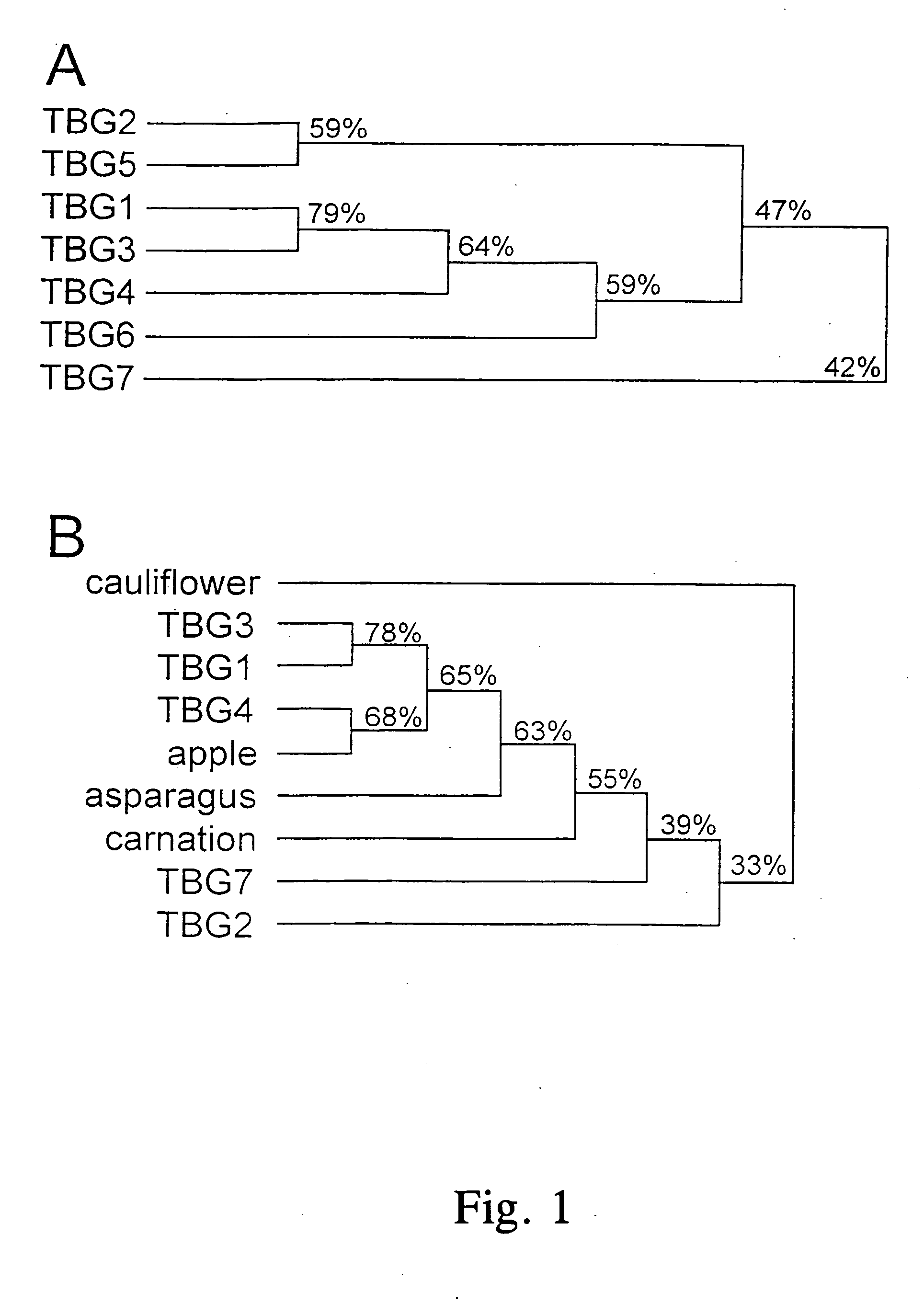
[0083] Degenerate primers were designed based on the highest shared deduced amino acid sequence identity we found between an apple (accession number P48980), asparagus (P45582) and carnation (Q00662) β-galactosidase cDNA clones. The two primers used for the first reaction were BG5′E1 (WSNGGNWSNATHCAYTAYCC) and BG3′E (CCRTAYTCRTCNADNGGNGG). A second reaction was done on the products of the first reaction using BG5′I1 (ATHCARACNTAYGTNTTYTGG) and BG3′E. The degeneracy code for the primer sequences is N=a+t+c+g; H=a+t+c; B=t+c+g; D=a+t+g; V=a+c+g; R=a+g; Y=c+t; M=a+c; K=t+g; S=c+g; and W=a+t. The 5′ and 3′ primers corresponded to amino acids 72-78 and 321-315 of the apple clone, respectively. Amplification was done using AmpliTaq DNA polymerase (Perkin Elmer, Norwalk, Conn.) and standard PCR conditions using the cDNA made for the first cDNA library described below as a template (Ausubel et al. 1987. In: Current Protocols in Molecular Biology, John Wiley and Sons, New York, N.Y.)....
example 3
cDNA Library Construction
[0084] Two cDNA libraries were constructed. The first comprised poly(A) RNA isolated from breaker, turning and pink fruit pericarp from ‘Rutgers’ plants. The cDNA synthesis and library construction was done exactly according to the manufacturers instructions for the ZAP-cDNA Gigapack II Gold Cloning Kit (Stratagene), the complete disclosure of which is fully incorporated herein by reference. First-strand cDNA synthesis was primed using a poly(dT) primer and inserts were directionally cloned into the Uni-Zap XR vector using EcoRI and XhoI restriction sites. The second library comprised poly(A) RNA isolated from all fruit tissues (except seeds) from immature green, mature green, breaker, turning, pink, red-ripe and over-ripe fruit of ‘Rutgers’ plants. The cDNA synthesis and library construction was done exactly according to the manufacturers instructions for the SuperScript Lambda System for cDNA synthesis and Cloning (Gibco BRL, Gaithersburg, Md.). First-str...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Gene expression profile | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com