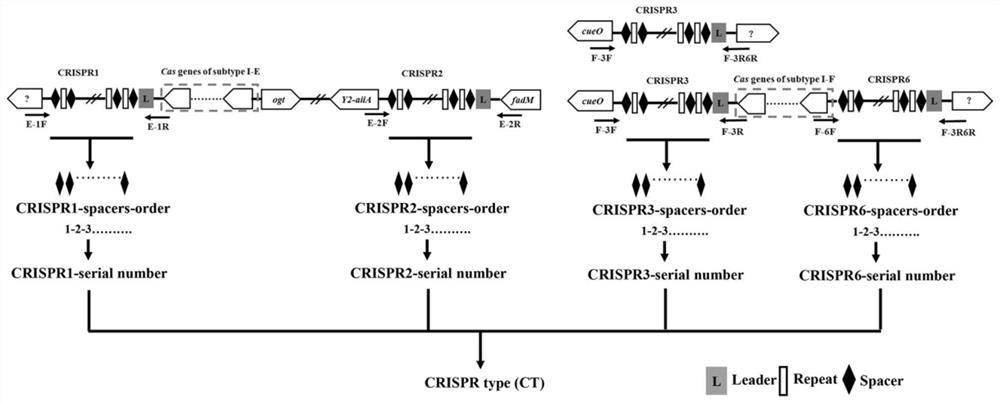
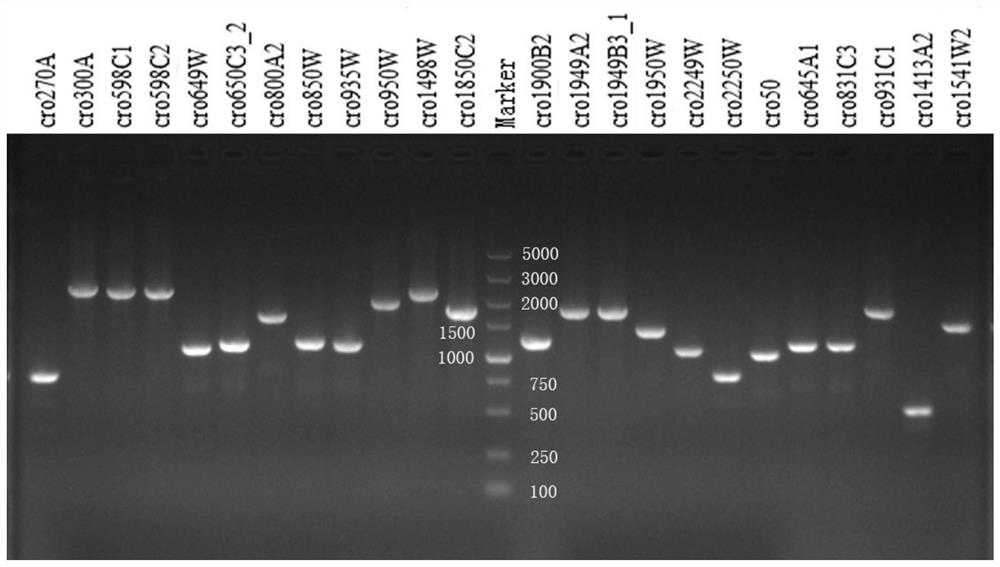
A CRISPR typing method for Cronobacter malonate
A technology of Cronobacter malonate and typing method, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problem of high cost of genome sequencing, and achieve low typing cost and good resolution The effect of high rate and high resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 164
[0033] Example 1 CRISPR typing method of 64 strains of Cronobacter malonate
[0034] (1) strain
[0035] The 64 strains used in this experiment were isolated from milk powder, edible fungus and vegetable samples in various regions of my country. Traditional biochemical identification and molecular detection have been completed, and they were identified as Cronobacter malonate.
[0036] (2) Bacterial DNA extraction
[0037] Genomic DNA of the above-mentioned 64 strains to be tested was extracted using the bacterial genomic DNA extraction kit from Magen Company, and the extracted DNA was stored at -20°C.
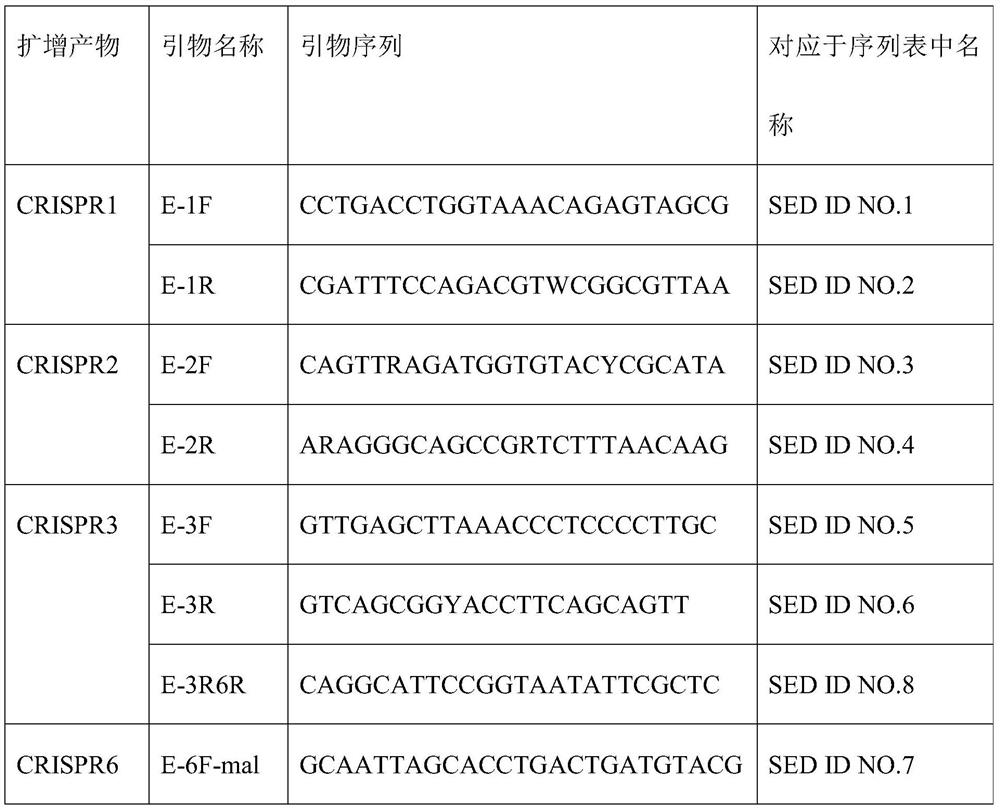
[0038] (3) Primer synthesis
[0039] Table 1 Primer list for CRISPR molecular amplification
[0040]
[0041]
[0042] The concentration of primers used in PCR amplification was 10 μmol / L.
[0043] (4) PCR amplification
[0044] 4.1 CRISPR1 amplification PCR reaction system:
[0045]
[0046] The PCR reaction conditions for CRISPR1 amplification were as follows:...
Embodiment 2
[0069] Example 2 CRISPR typing of Cronobacter malonate and comparison of the resolution of MLST typing methods
[0070] (1) MLST classification
[0071] Seven housekeeping genes of Cronobacter malonionum were mainly selected for amplification and sequencing, and the obtained sequence information was uploaded to a special website ( https: / / pubmlst.org / cronobacter / ) to obtain the corresponding MLST type, see the MLST website for specific experimental operation methods.
[0072] (2) Comparison of the resolution of the two typing methods
[0073] Table 3 Comparison of MLST typing and CRISPR typing results of Cronobacter malonobacter
[0074]
[0075]
[0076]
[0077] The resolution evaluation is quantified by the Simpson diversity index (D), and its calculation formula is: D=1-∑[nj(nj-1)] / [N(N-1)], where nj represents the value of the jth band type The number of strains, N represents the total number of experimental strains. The larger the D value, the stronger the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com