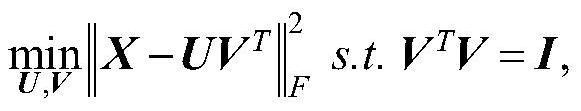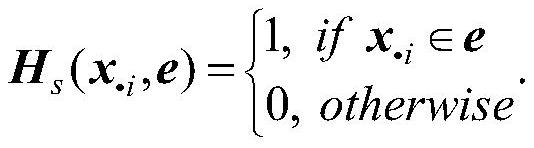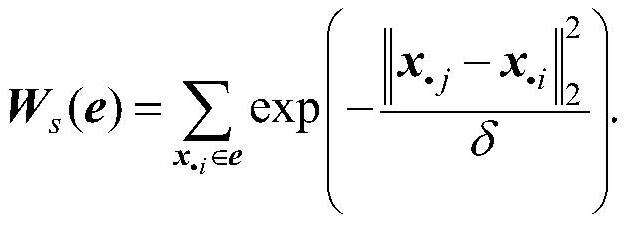Double clustering method for tumor gene expression profile data based on double hypergraph regularization
A tumor gene and expression profile technology, applied in the field of double clustering of tumor gene expression profile data, can solve the problem that the principal component analysis method cannot mine the inherent geometric structure of the data, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0059] The present invention will be further described below.
[0060] Concrete steps of the present invention are:
[0061] Step Ⅰ: Decompose the tumor gene expression profile data into a gene clustering matrix and a sample clustering matrix by principal component analysis;
[0062] Step II: Construct a sample hypergraph based on samples of tumor gene expression profile data;
[0063] Step Ⅲ: Construct a gene hypergraph according to the genes of the tumor gene expression profile data;
[0064] Step Ⅳ: Using the sample hypergraph and the gene hypergraph as the sample hypergraph regularization item and the gene hypergraph regularization term respectively as principal component analysis, determine the form of the optimization objective function;
[0065] Step V: optimize the sample clustering matrix and gene clustering matrix in step I by optimizing the objective function, and obtain the optimized sample clustering matrix and gene clustering matrix;
[0066] Step VII: realize...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com