Hla-restricted epitopes encoded by somatically mutated genes
A cell and cell receptor technology, applied in receptor/cell surface antigen/cell surface determinant, translation product of oncogene, anti-receptor/cell surface antigen/cell surface determinant immunoglobulin, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Materials and methods
[0048] cell line. T2 cells (ATCC, Manassas, VA) were incubated at 37°C in 5% CO 2 were cultured in the presence of 10% FBS (GEHyclone, Logan, Utah, USA), 1% penicillin streptomycin (Life Technologies) and 20IU / mL recombinant human IL-2 (Proleukin TM , Prometheus Laboratories) in RPMI-1640 (ATCC). T2A3 cells (a gift from Eric Lutz and Liz Jaffee, Johns Hopkins University (JHU)) were grown under the same conditions as T2 cells, but with the addition of 500ug / mL Geneticin (Life Technologies) and 1 × Non-essential amino acids (Life Technologies).
[0049] Phage display library construction. Oligonucleotides were synthesized at DNA 2.0 (Menlo Park, CA) using mixed and split pooldegenerate oligonucleotide synthesis. Oligonucleotides were incorporated into pADL-10b phagemid (Antibody Design Labs, San Diego, CA). The phagemid contains an F1 origin, and a transcriptional repressor unit consisting of a lactose operator and a lactose repressor to limi...
Embodiment 2
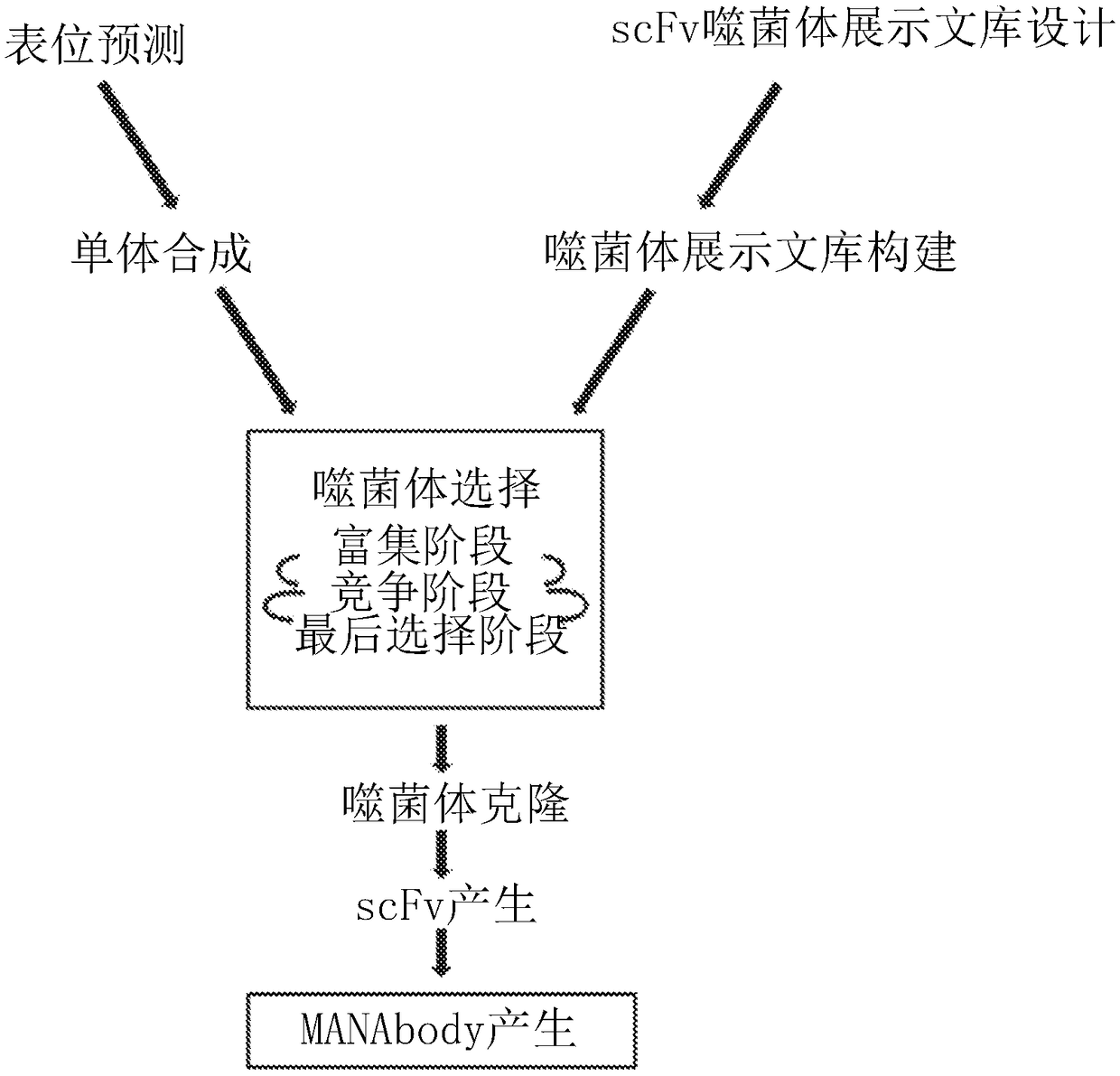
[0073] Design and construction of scFv-based phage display scFv libraries. The inventors initiated these studies in an attempt to generate antibodies against the mutant KRAS peptide in mice (the basis for this selection is described below). Using conventional methods to obtain monoclonal antibodies after immunization of mice, these efforts failed because no antibodies specifically reactive with MANA were identified. The inventors therefore turned to a phage display method for generating MANAbodies ( figure 1 ). The design of phage display libraries follows principles used in published studies (22) and includes some specific features. The framework of the library is based on the scFv sequence of the humanized 4D5 antibody (Trastuzumab) raised against the protein encoded by ERBB2 (23). This framework was chosen for its stability on phage and its ease of conversion into soluble scFv, Fab or antibody (22, 24). A high-resolution crystal structure of humanized 4D5 has identified...
Embodiment 3
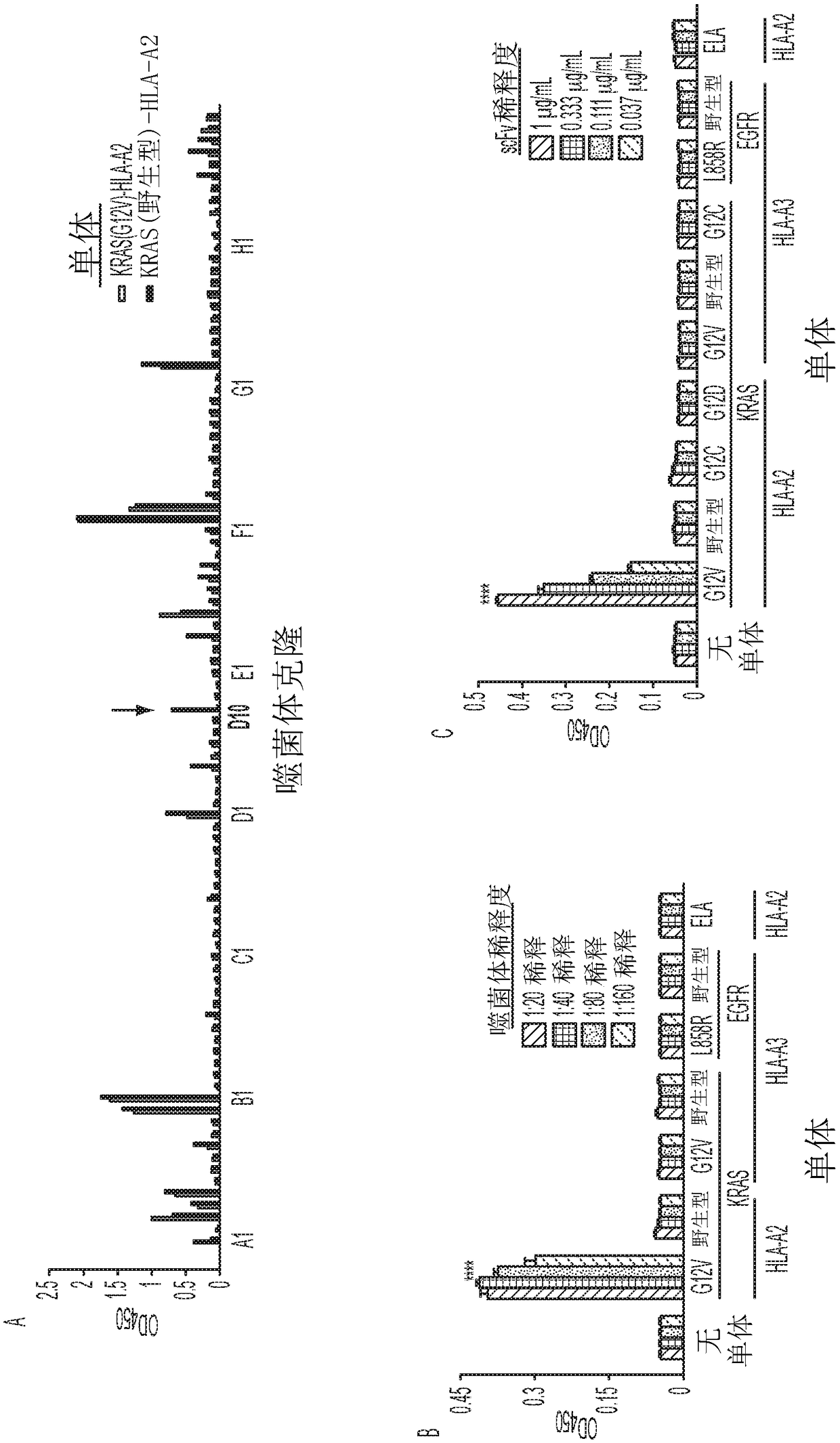
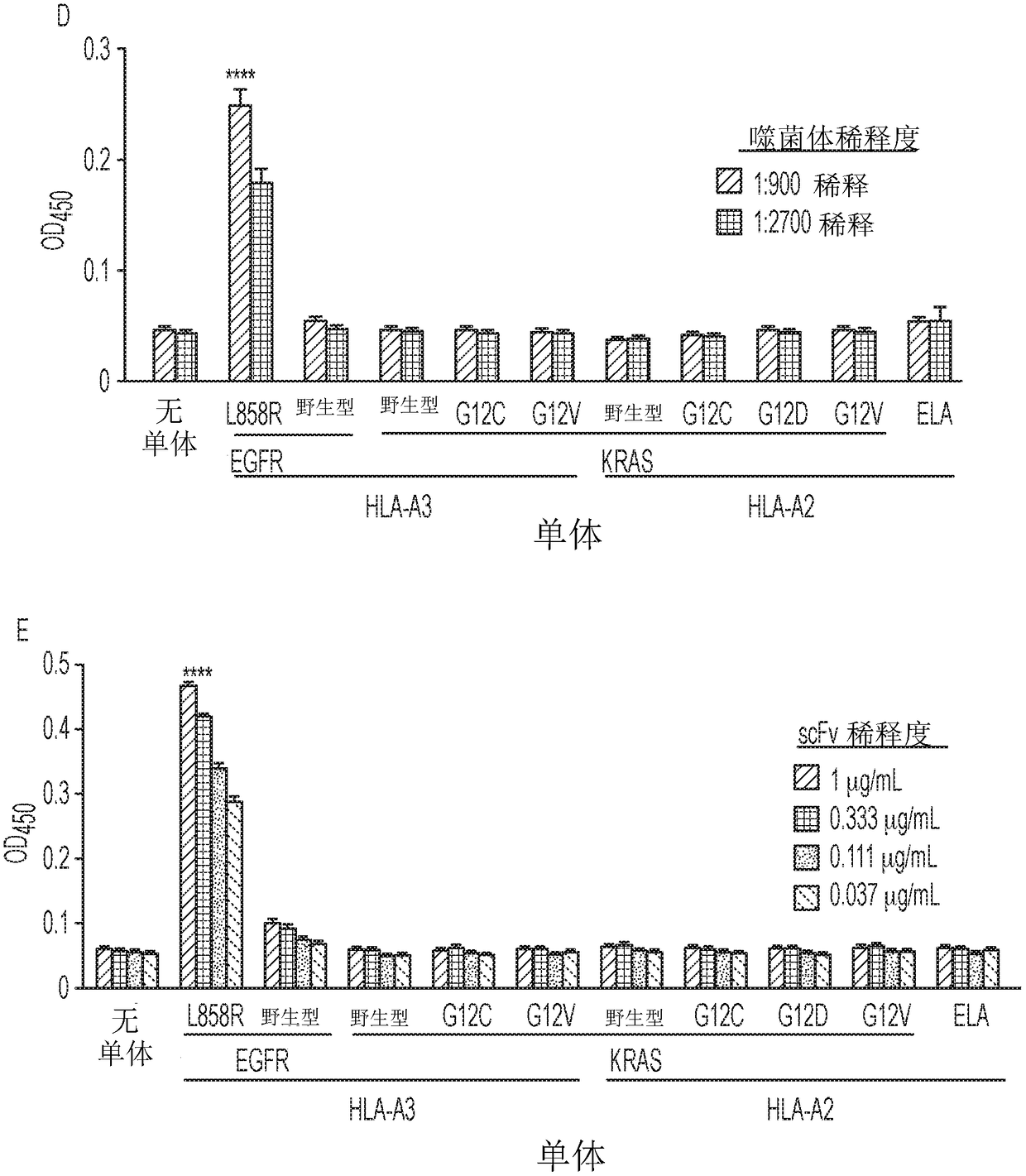
[0076] Target selection and competition strategies for identifying selectively responsive phage clones. The inventors selected MANAbody targets based on the frequency of specific mutations and their predicted strength of binding to HLA alleles. KRAS is one of the most frequently mutated genes in human cancers, with mutations particularly prevalent in pancreatic, colorectal and lung adenocarcinomas. The inventors chose the G12V mutation as a target because a related peptide containing it is predicted to bind with high affinity to HLA-A2, the most common HLA allele in many human populations (32). This bioinformatics (in silico) prediction was performed using the NetMHC v3.4 algorithm (33-35). Additionally, based on structural studies of other peptide-HLA complexes, a key mutated residue (V at codon 12) was expected to be exposed on the surface of HLA proteins (36). The peptide KLVVVGAVGV (SEQ ID NO: 4), in which the valine residue (V) at position 8 represents the G12V mutation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com