Biological species identification method based on genetic barcode
An identification method and barcode technology, applied in the direction of instruments, calculations, electrical digital data processing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Example 1: Identification of Campylobacter jejuni subspecies NCTC11168 bacillus
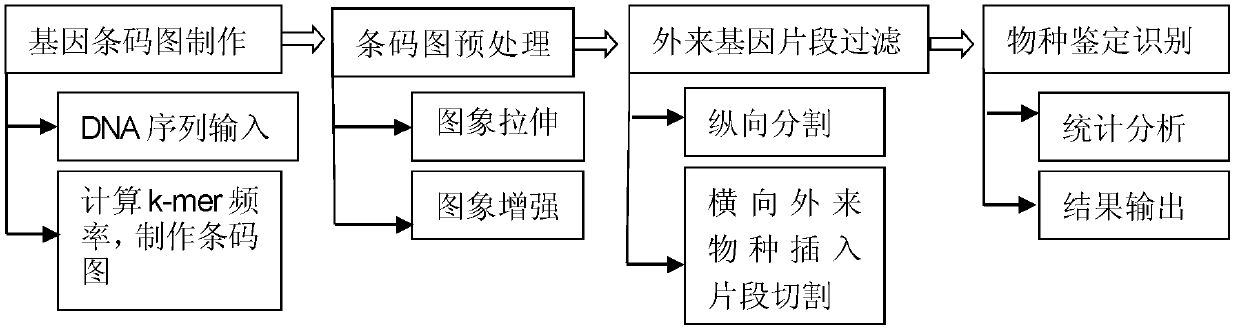
[0105] We identify the subspecies NCTC11168 of the Campylobacter jejuni family. According to the method proposed in the present invention, the gene barcode image of the Campylobacter jejuni subspecies NCTC11168 bacillus is produced, preprocessed, vertically segmented, horizontally retrieved, and cut the gene fragment of the foreign insertion species , and finally calculate the acquaintance with the species in the prokaryotic database, and obtain the similarity measure of Campylobacter jejuni subsp. Table 3 shows:
[0106] Table 2: Similarity measures between NCTC11168 bacillus and the same family of bacteria
[0107] Campylobacter jejuni subsp. NCTC11168
[0108] Table 3: Similarity measure between NCTC11168 bacillus and heterophyletic bacteria
[0109] Hidden Acidophilus JF-5
[0110] It can be seen from Table 2 that the similarity measure between Campylobacter jejun...
Embodiment 2
[0111] Example 2: Identification of Chlamydia trachomatis D / UW-3 / CX
[0112] We selected Chlamydia trachomatis D / UW-3 / CX to do the second group of species identification and recognition experiments, and made and processed the gene barcode image of Chlamydia trachomatis D / UW-3 / CX according to the method proposed in the present invention , and finally calculate the acquaintance with the species in the prokaryotic database, and get the similarity measure between Chlamydia trachomatis D / UW-3 / CX and the same family of bacteria as shown in Table 4. Chlamydia trachomatis D / UW-3 / CX and The similarity measures of other alien bacteria are shown in Table 5:
[0113] Table 4: Similarity measure of Chlamydia trachomatis D / UW-3 / CX and the same family of bacteria
[0114] Chlamydia trachomatis D / UW-3 / CX
[0115] Table 5: Similarity measure between Chlamydia trachomatis D / UW-3 / CX and heterophyletic bacteria
[0116] Hidden Acidophilus JF-5
[0117] It can be seen from Ta...
Embodiment 3
[0118] Example 3: Haemophilus influenzae Rd KW20
[0119] We selected Haemophilus influenzae Rd KW20 to do the third group of species identification and recognition experiments. According to the method proposed by the present invention, the gene barcode image of Haemophilus influenzae Rd KW20 was produced, processed, and finally calculated with the species in the prokaryotic database Similarity, the similarity measure of Haemophilus influenzae Rd KW20 and the same family of bacteria is shown in Table 4, and the similarity measure of other heterogeneous bacteria is shown in Table 5:
[0120] Table 6: Similarity measure between Haemophilus influenzae Rd KW20 and the same family of bacteria
[0121] Haemophilus influenzae Rd KW20
[0122] Table 7: Similarity measures between Haemophilus influenzae Rd KW20 and heterogeneous bacteria
[0123] Thermotoga maritima MSB
[0124] Escherichia coli K12
[0125] It can be seen from Table 6 that the similarity...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com