Hemopexin-like structure as polypeptide-scaffold
A target molecule and specific technology, which is used in the field of preparing polypeptides with specific binding properties to predetermined target molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[1278] Example 1 Overlap extension ligation PCR (OEL-PCR)
[1279] Linear expression elements are assembled in a modular fashion by a two-step PCR protocol utilizing the principle of overlapping DNA ligation. In standard PWO-PCR, the intronless open reading frame is amplified with sequence-specific end-bridging primers, generating homologous sequences that overlap the flanking DNA sequences. 2 μl of the first PCR mix containing approximately 50 ng of the extended gene fragment (gene module) was transferred to the second PWO-PCR mix. To this mixture was provided 50ng to 100ng of pre-generated DNA fragments (promoter module and terminator module) and the respective sequence-specific end primers at 1 μM each. Typically, the second PCR step contains 30 cycles. The physical parameters of the PCR properties are adjusted according to the needs of the DNA fragments to be ligated.
Embodiment 2
[1280] Embodiment 2 synthetic PEX2 DNA library
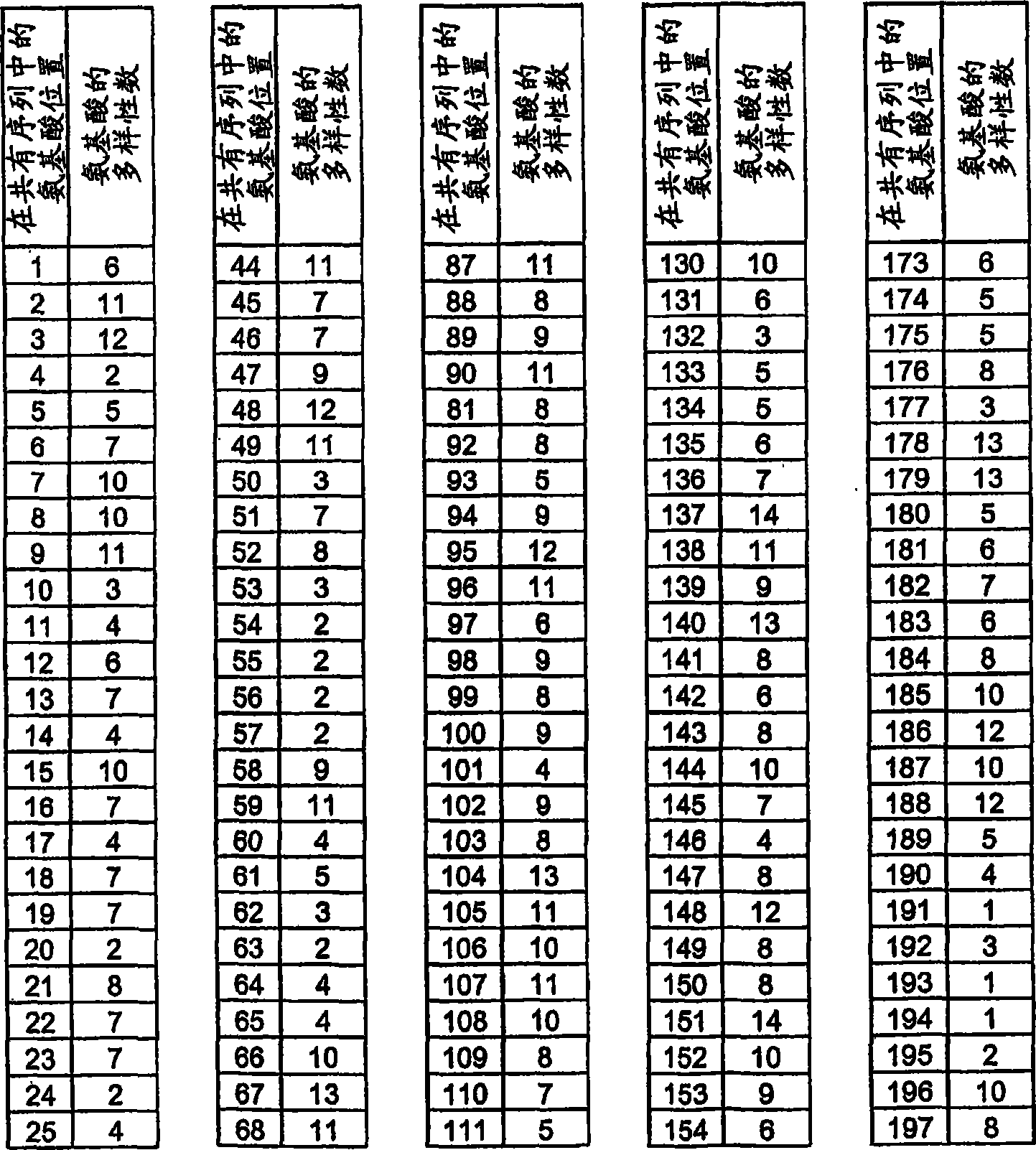
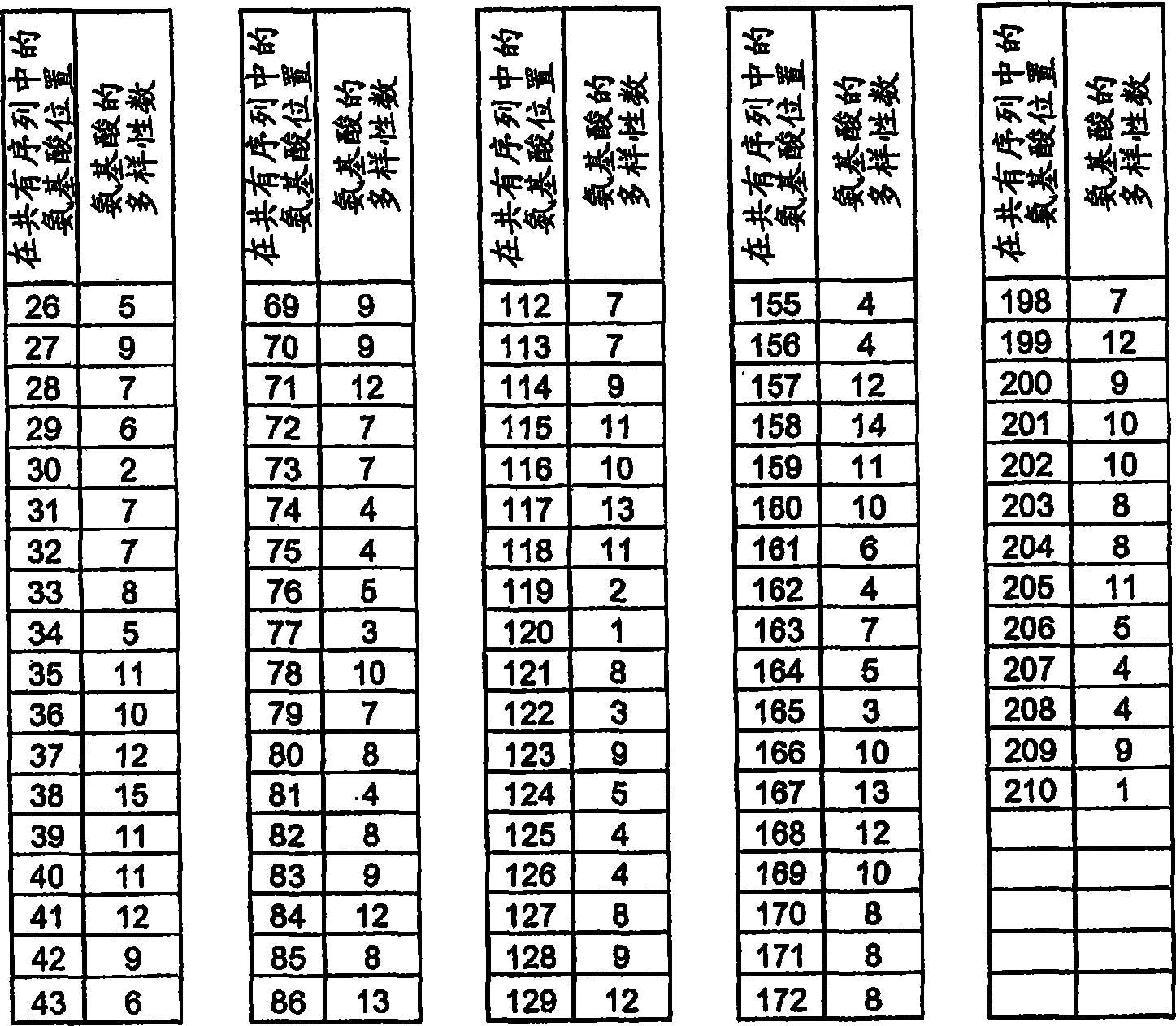
[1281] Amino acids 64, 65, 86, 112, 113, 114, 130, and 132 encoding the hemopexin-like domain (equivalent to 528 (Gln), 529 (Glu), 550 (Arg ), 576(Lys), 577(Asn), 578(Lys), 594(VaI) and 596(Lys)) of the PEX2 triplet codons were randomized by the NNK-motif. The human wild-type PEX2 DNA sequence is divided into three sequence parts. Standard PWO-PCR to which 10 ng of vector template pIVEX2.1MCS PEX2 and 1 μM each of primers PEX2forw (SEQ ID NO: 63) and PEXR4 (SEQ ID NO: 64) were provided amplified a 1 bp-218 bp fragment. The 402bp-605bp fragment was amplified in standard PWO-PCR with 10 ng vector template pIVEX2.1MCS PEX2 and 1 μM each of primers PEXF4 (SEQ ID NO: 65) and PEX2rev (SEQ ID NO: 66). The formed sequence 196bp-432bp overlaps with the DNA fragment 1bp-218bp and 402bp-605bp and is primer PEXF1 (SEQ ID NO: 67) and PEXR1 (SEQ ID NO: 68) each 1 μ M and PEXR3 (SEQ ID NO: 69), PEXR2 ( SEQ ID NO: 70) and PEXF2 (SEQ ID NO: 7...
Embodiment 3
[1282] Example 3 Cell-free protein transcription and translation in vitro
[1283] Linear expression elements were transcribed and translated in the RTS 100HY E. coli system according to the manufacturer's instructions. Linear DNA templates (100ng-500ng) were incubated at 30°C. Optionally 6 μl of GroE supplement (Roche) was added.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com