Optimum Design Method of Pathogen Species-Specific PCR Primers
A technology for optimizing design and primer design, applied in the field of bioinformatics analysis, can solve problems such as affecting the sensitivity and specificity of primer design, affecting the performance of multiplex PCR combination reactions, and being inapplicable, so as to avoid strain-specific problems.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The optimal development of embodiment 1 specific primer design method
[0060] The present invention has finally established and developed a specific primer design method through a large number of bioinformatics analysis and optimization experiments. The specific steps of the scheme are as follows:
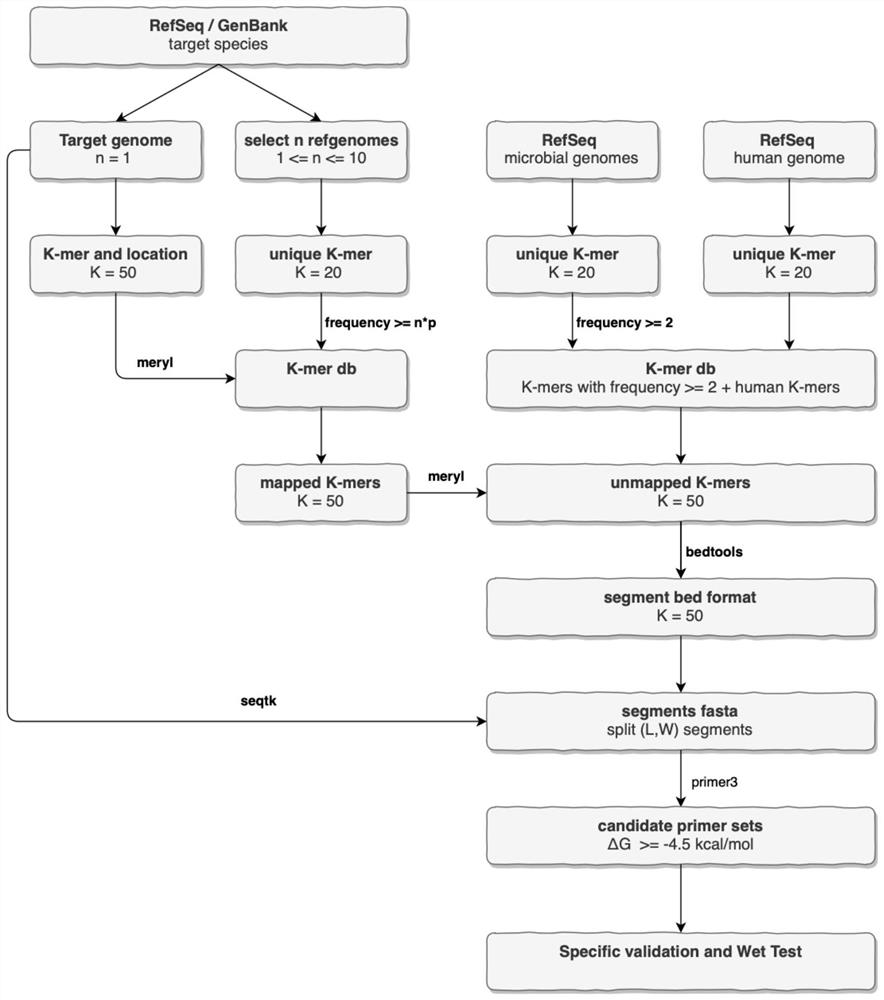
[0061] 1. Construction of the background comparison library:
[0062] 1. Obtain the reference genomes of all microbial species in NCBI RefSeq, and divide K-mer for each genome to form a unique K-mer set. The value of K is 18-20, preferably 20.
[0063] 2. Select the K-mer with a frequency >= 2 from the unique K-mer set at the microbial species level as the candidate K-mer set. The value of K is 18-20, preferably 20.
[0064] 3. The human genome is divided into unique K-mer collections according to method 1, and combined with the candidate K-mer collections at the microbial species level in method 2 to construct a K-mer db and obtain a background comparison library. The ...
Embodiment 2
[0076] Example 2 Bacteria / fungus / virus species-specific PCR primer design and evaluation
[0077] 1. Take the bacteria Mycoplasma pneumoniae / fungus Aspergillus flavus / virus EBV as examples to design PCR primers respectively:
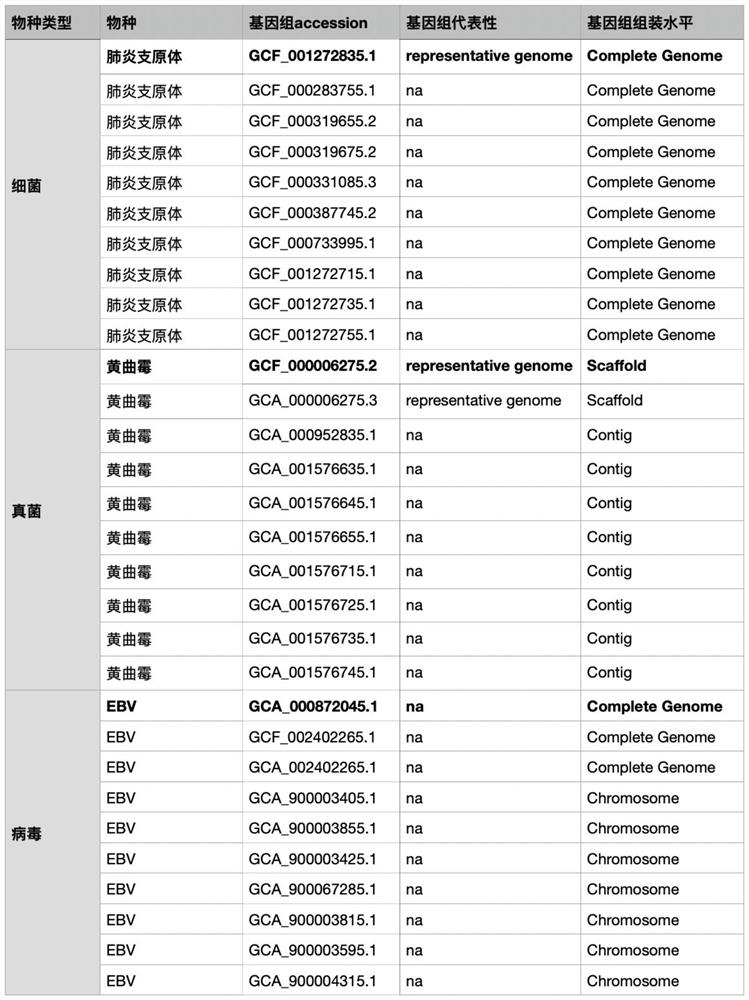
[0078] 1. Screen n target species genomes from the NCBI RefSeq / GenBank library. The value of n is 10, see figure 2 .
[0079] 2. Divide the screened genomes into K-mers to obtain unique K-mer sets. The value of K is 20.
[0080] 3. Use meryl software to build a K-mer db for the unique K-mer set in method 2, extract K-mers with a frequency greater than n*p from it, and use it as a K-mer set shared by the species level, and construct a species-shared K -mer comparison library. The value of K is 20, the value of n is 10, and the value of p is 0.8.
[0081] 4. Use the NCBI RefSeq / GenBank optimal reference genome screening rules to screen the reference genome of the target species, divide the K-mer, and retain the position information of all K-mers, an...
Embodiment 3
[0114] The wet test verification of embodiment 3 species-specific PCR primers
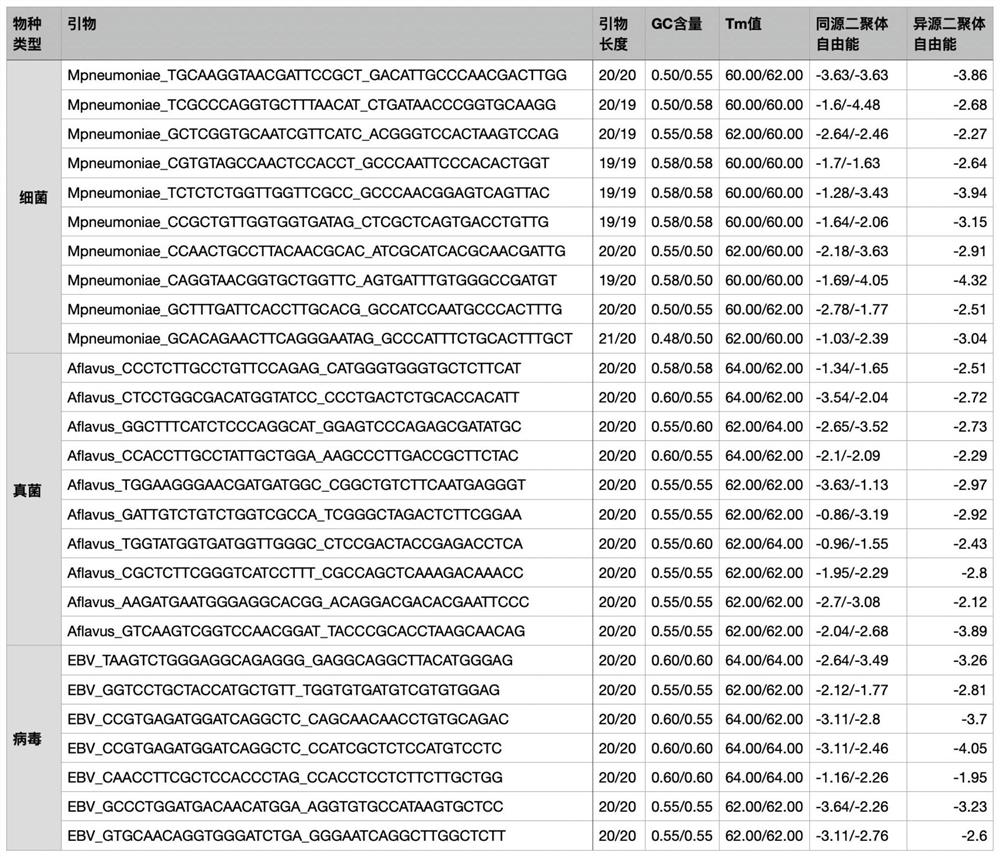
[0115] This example is further validated by the wet test of Aspergillus flavus species-specific PCR primers (F: 5'-CCCTCTTGCCTGTTCCAGAG-3' (SEQ ID NO.1), R: 5'-CATGGGTGGGTGCTCTTCAT-3' (SEQ ID NO.2)) As an example, to illustrate the effectiveness of the specific primers designed based on the ideas of the present invention.
[0116] 1) Reagent consumables
[0117] Enzyme-free sterile water: ThermoFisher, Nuclease-Free Water (not DEPC-Treated) (Cat. No.: AM9937); Qubit Fluorometer DNA Detection Kit: Qubit 1X dsDNA HS Assay Kit (Cat. No.: Q33231); PCR amplification enzyme: GXL DNA Polymerase (R050A).
[0118] 2) Primer verification
[0119] The nucleic acid extracted from the Aspergillus flavus standard was used as a template for PCR reaction verification, and at the same time, gDNA and Zymo bacteria DNA templates were added in parallel to the PCR reaction system to simulate real clinical samples f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com