Porcine epidemic diarrhea virus m protein affinity peptide and its screening method
A porcine epidemic diarrhea and affinity peptide technology, applied in the direction of peptides, can solve the problems of the world's pig industry loss, vaccination protection failure, etc., and achieve the effect of inhibiting virus replication and good antiviral effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Prokaryotic expression of recombinant plasmid pET32a-M:
[0036] According to the published PEDV M gene sequence in GenBank, design primers P1 and P2:
[0037] P1: 5'-CGC GAATTC GCCATGTCTAACGGTTCTAT-3' (the underline indicates that the introduced restriction site is EcoRI)
[0038] P2: 5'-CGC CTCGAG TTAGACTAAATGAAGCAC-3' (the underline indicates that the introduced restriction site is Xho I)
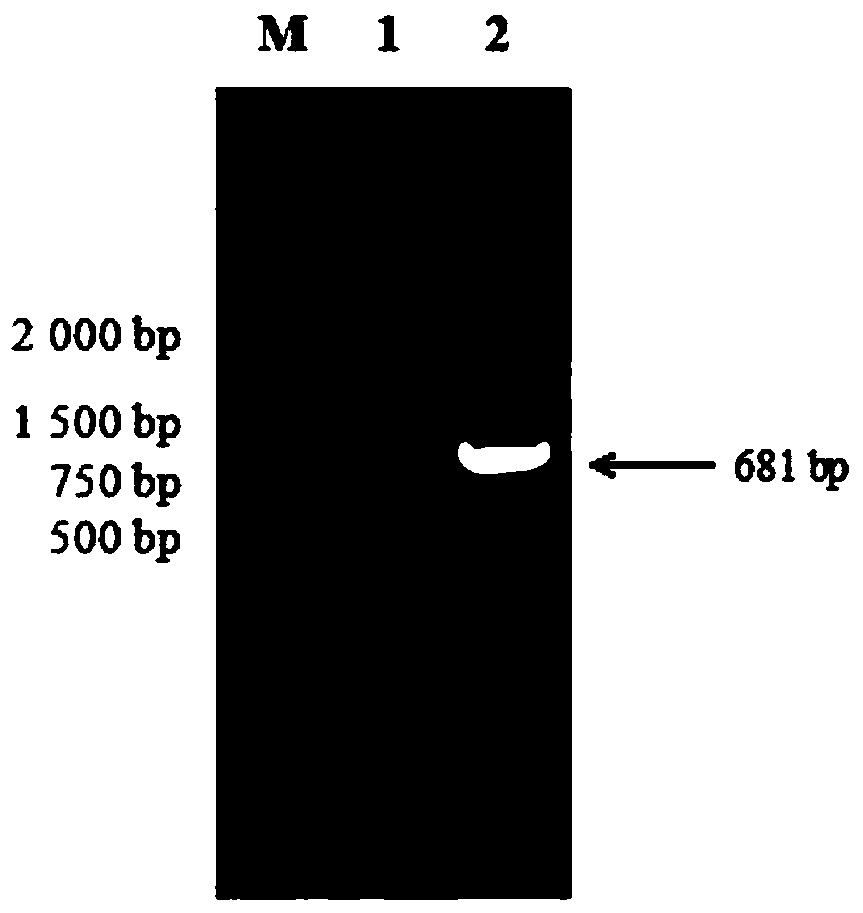
[0039] The standard virus strain CV777 was selected, genomic DNA was extracted, and the PEDVM gene fragment was amplified by PCR. Amplification conditions: pre-denaturation at 94°C for 10 minutes, denaturation at 94°C for 30 seconds, annealing at 50.2°C for 30 seconds, extension at 72°C for 1 minute, and pre-denaturation at 94°C for 10 minutes, 94°C Denaturation at ℃ for 30s, annealing at 49.6℃ for 30s, extension at 72℃ for 1min, a total of 30 cycles, and a final extension at 72℃ for 10min. The PCR product was analyzed by agarose gel electrophoresis with a mass fraction of 1...
Embodiment 2
[0041] Preparation of PEDV M polyclonal antibody:
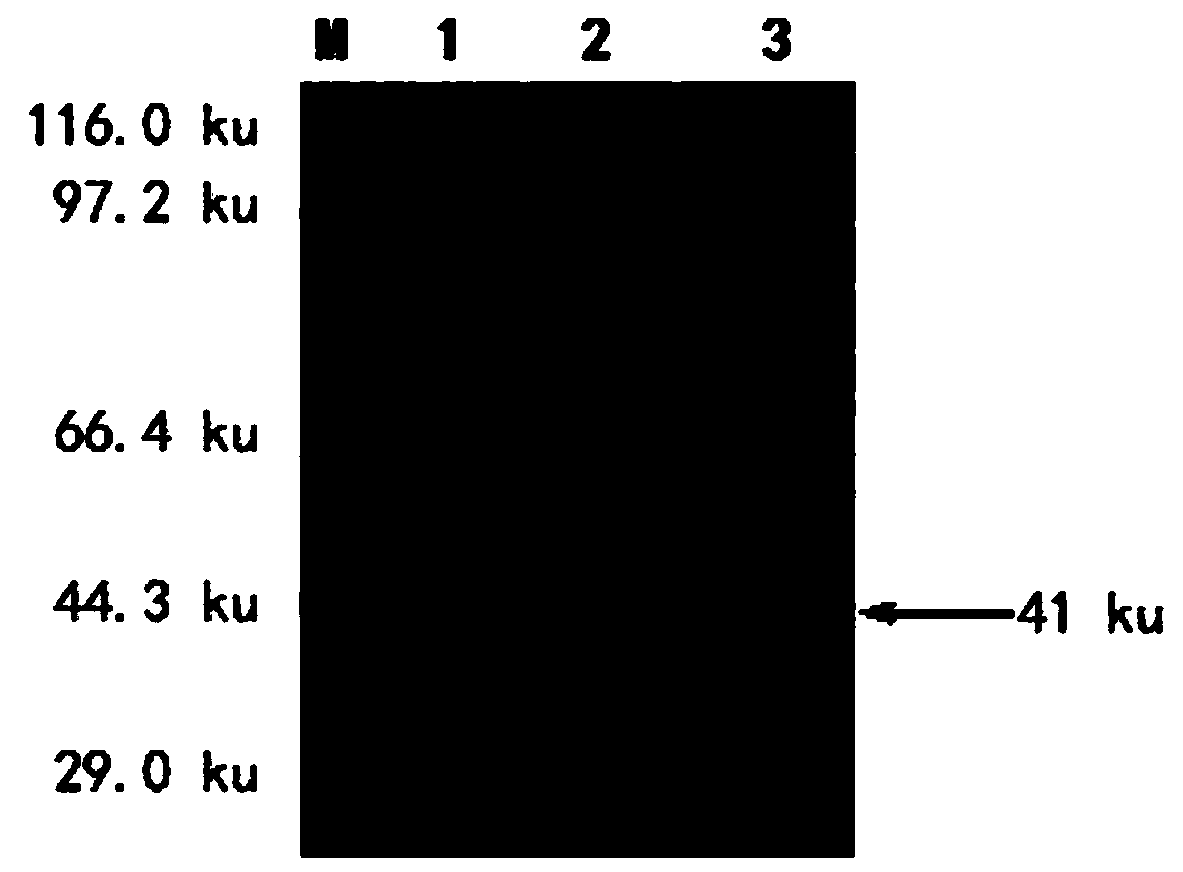
[0042] Select 2-3kg New Zealand white rabbits, the first injection is the purified recombinant M protein and an equal volume of Freund's complete adjuvant after complete emulsification, multi-point subcutaneous injection on the back, the dose is 2 mg / rat, and then the Freund's incomplete adjuvant and equal volume The purified recombinant M protein was completely emulsified and immunized once at intervals of 1 week. After 3 immunizations, the purified recombinant M protein without adjuvant was used to boost the immunization. After 7 days, blood was collected from the rabbit heart to obtain serum, and anti-PEDV M protein polyclonal antibody was obtained. Indirect ELISA was used to detect antibody titers, and Western blot was used to identify the characteristics of polyclonal antibodies. The results of SDS-PAGE analysis showed that a specific band appeared at 41ku after the induction of the recombinant plasmid bacterial liquid, ...
Embodiment 3
[0044] Screening and functional determination of PEDV M affinity peptide:
[0045] (1) Coating of M protein
[0046] The M protein with a concentration of 15 μg / 100 μL was coated on an ELISA plate, blocked overnight at 4°C, and washed 6 times with 0.1% TBST. After washing, add 100 μL of the diluted phage library, shake for 10 minutes, stop for 10 minutes, a total of 40 minutes, wash 10 times with 0.1% TBST, add 100 μL of eluent, shake for 30 minutes, add the supernatant to the EP tube, add 15 μL Neutralizing solution, phage titer determination and amplification, respectively, and subsequent screening. Carry out the next three rounds of screening according to the above steps, and gradually increase the selection pressure, that is, the coating concentration of M protein in each round will be halved sequentially, and the number of TBST washings will be increased by 5 times in each round.
[0047] (2) Amplification of affinity phages: The positive phages after each screening wer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com