Fluorescent quantitative PCR method for detecting toxigenic chlamydia pneumoniae and corresponding kit
A Chlamydia pneumoniae, fluorescent quantitative technology, applied in the direction of microorganism-based methods, biochemical equipment and methods, microbial measurement/inspection, etc., can solve the difficult to distinguish bacterial, viral Mycoplasma pneumoniae or Chlamydia pneumoniae infection, test To solve problems such as complicated process and difficult clinical medication, to achieve stable test results, wide application range, rigorous and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
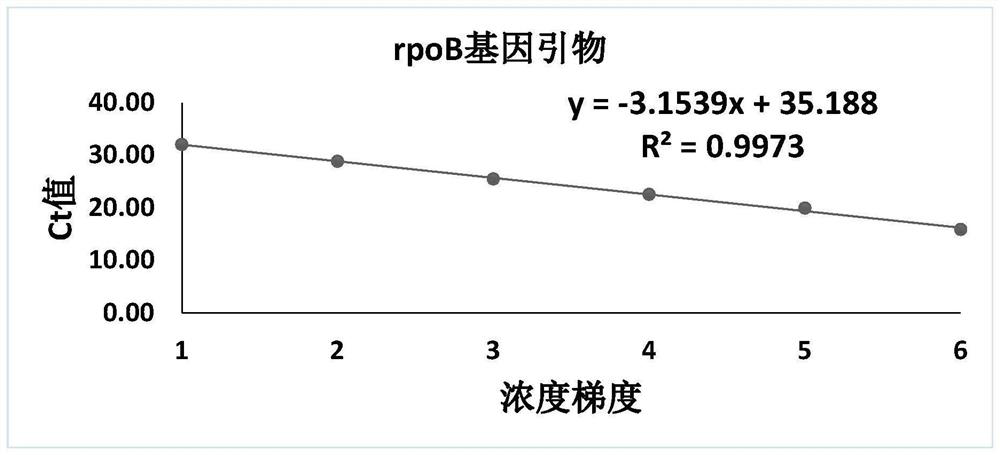
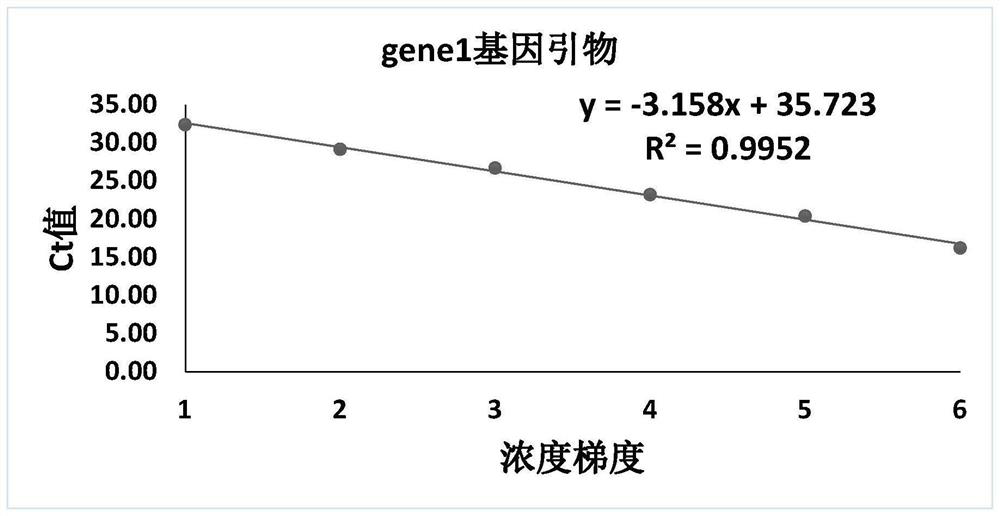
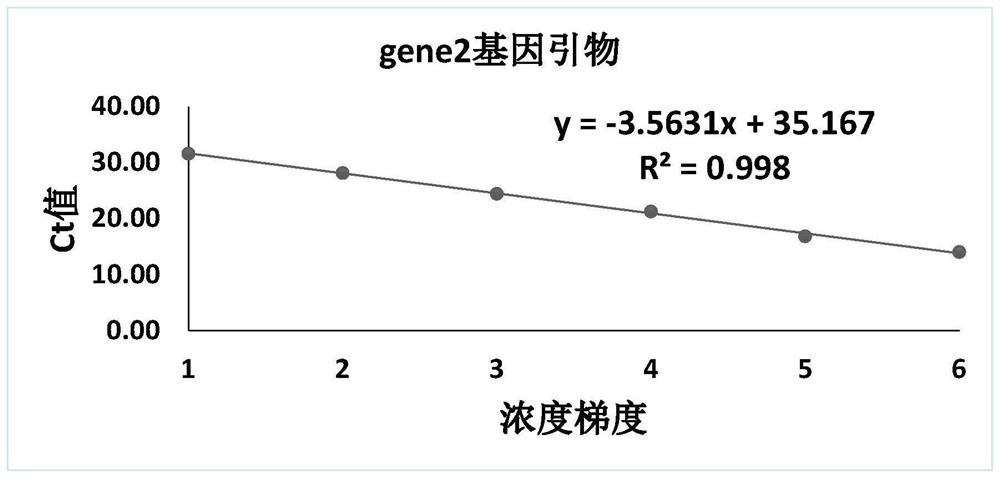
[0048] Embodiment 1 detection primer amplification standard curve
[0049] 1. Microbial culture
[0050] Alkaline peptone water is used as the culture medium for toxigenic Vibrio vulnificus, and it grows well in alkaline peptone water or plates with a pH of 8.8-9.0. The diameter of the colony on the alkaline plate is 2mm, round, smooth and transparent. The vigorously growing microbial samples were selected for subsequent experiments.
[0051] 2. Genomic DNA Extraction
[0052] Genomic DNA was extracted according to the instructions of Qiagen's QIAamp DNA Mini Kit.
[0053] 3. Production of standard plasmids for amplified products
[0054] Use the DNA extracted from Chlamydia pneumoniae to amplify with the primers of rpoB gene, gene1 gene, gene2 gene and gene4 gene respectively. After competent cells, the plasmid is amplified on a large scale. The primer sequences of the five genes are shown in SEQ ID NO: 1-10, and the details are shown in Table 2.
[0055] Table 2
[0...
Embodiment 2
[0067] Embodiment 2 Positive and negative sample detection situation
[0068] 1. Microbial culture
[0069] With embodiment 1.
[0070] 2. Genomic DNA Extraction
[0071] With embodiment 1.
[0072] 3. qPCR detection of 5 target genes
[0073] According to the instructions of BioRad iQ SYBR Green SuperMix, the qPCR amplification of the sample was carried out, and the Ct value reading corresponding to each pair of primers was obtained. Each qPCR reaction was set up with 3 biological repeats and 3 technical repeats, and the qPCR reaction system and amplification reaction conditions are shown in Table 5.
[0074] table 5
[0075]
[0076]
[0077] 4. Result Analysis
[0078] The qPCR amplification results of 5 pairs of primers showed that Chlamydia pneumoniae showed positive results of one or more genes of rpoB gene, gene1 gene, gene2 gene and gene3 gene, while other Chlamydia species showed negative results. Therefore, for Chlamydia pneumoniae and other Chlamydia ge...
Embodiment 3
[0082] The sensitivity of embodiment 3 detection system
[0083] 1. Microbial culture
[0084] Use alkaline peptone water as the culture medium for Vibrio vulnificus, and grow well in alkaline peptone water or plates with a pH of 8.8-9.0. The diameter of the colony on the alkaline plate is 2mm, round, smooth and transparent. Select vigorously growing microbial samples for subsequent experiments, collect the bacteria and calculate the concentration, and dilute by 10 to get 10 bacteria / ml, 100 bacteria / ml, 1000 bacteria / ml, and 10000 bacteria / ml. Bacterial solutions with different concentration gradients were used for subsequent DNA extraction experiments.
[0085] 2. Genomic DNA Extraction
[0086] With embodiment 1.
[0087] 3. qPCR detection of 5 target genes
[0088] With embodiment 2.
[0089] 4. Result Analysis
[0090] The qPCR amplification results of 5 pairs of primers showed that within the range from 10 bacteria to 10,000 bacteria, Chlamydia pneumoniae showed p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com