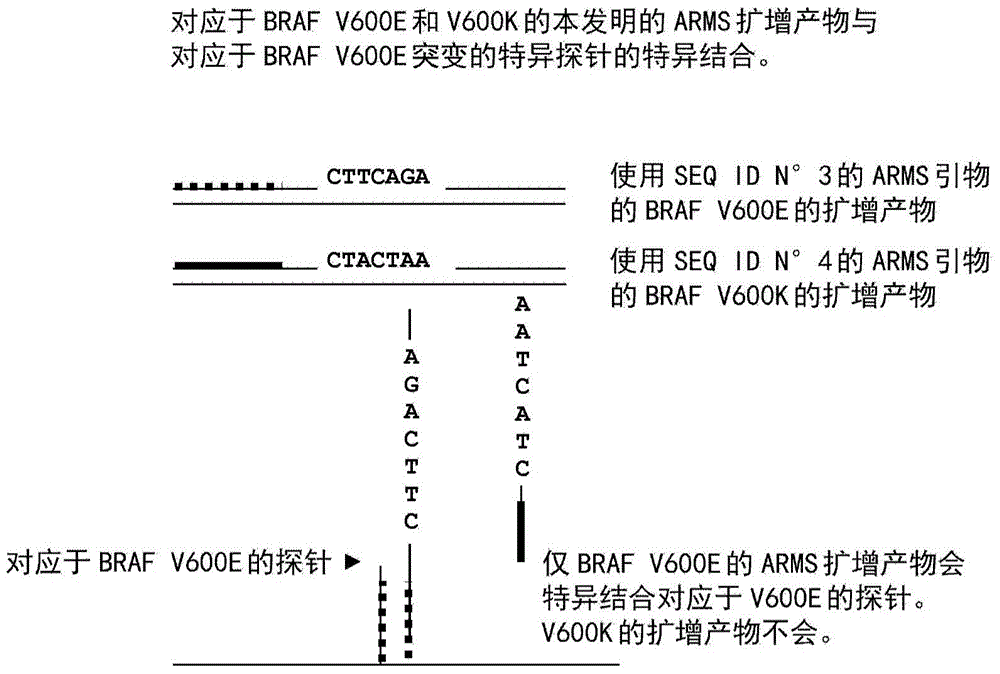
Method for detection of BRAF and PI 3K mutations
A test sample, V600K technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of low abundance, inability to accurately detect low abundance mutations, failure to pass, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0186] Prepare the following reagent mix for multiplex ARMS amplification of BRAF mutations V600E and V600K in samples / cell lines / clones:
[0187]
[0188] Next, 30 μl of 5 μl of total eluate obtained from paraffin-embedded tissue sections (samples), or from different cell lines or clones, was added to a final reaction volume of 50 μl.
[0189] The thermocycling conditions for PCR are:
[0190]
[0191]
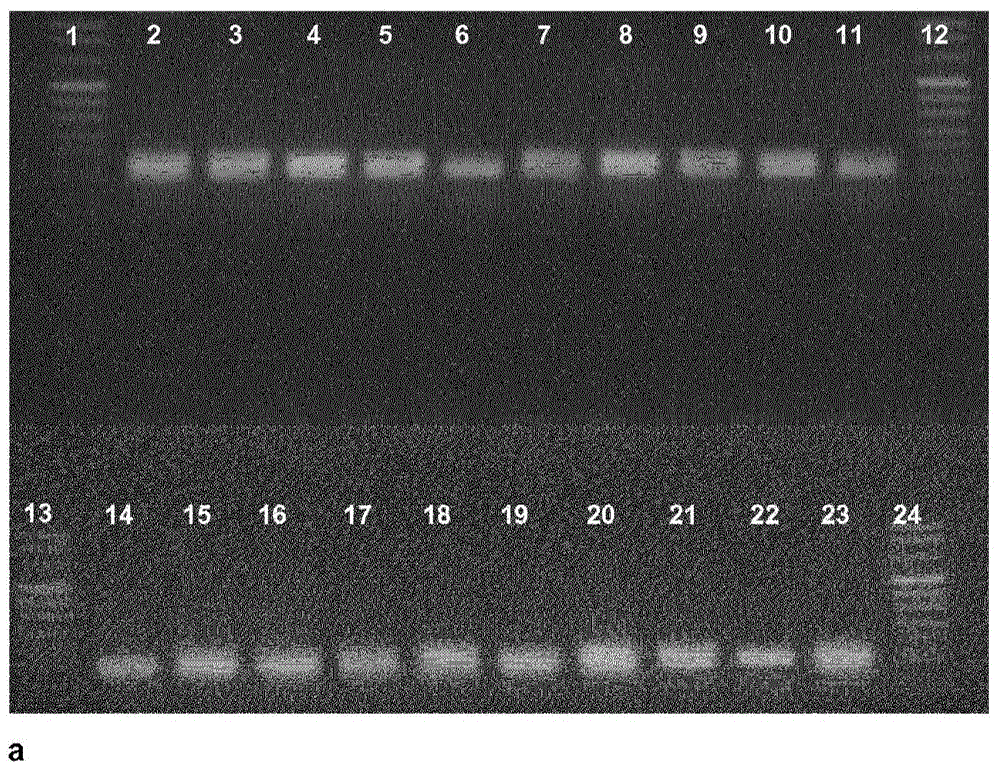
[0192] The results of the BRAF ARMS multiplex amplification are shown in figure 2 and image 3 middle.
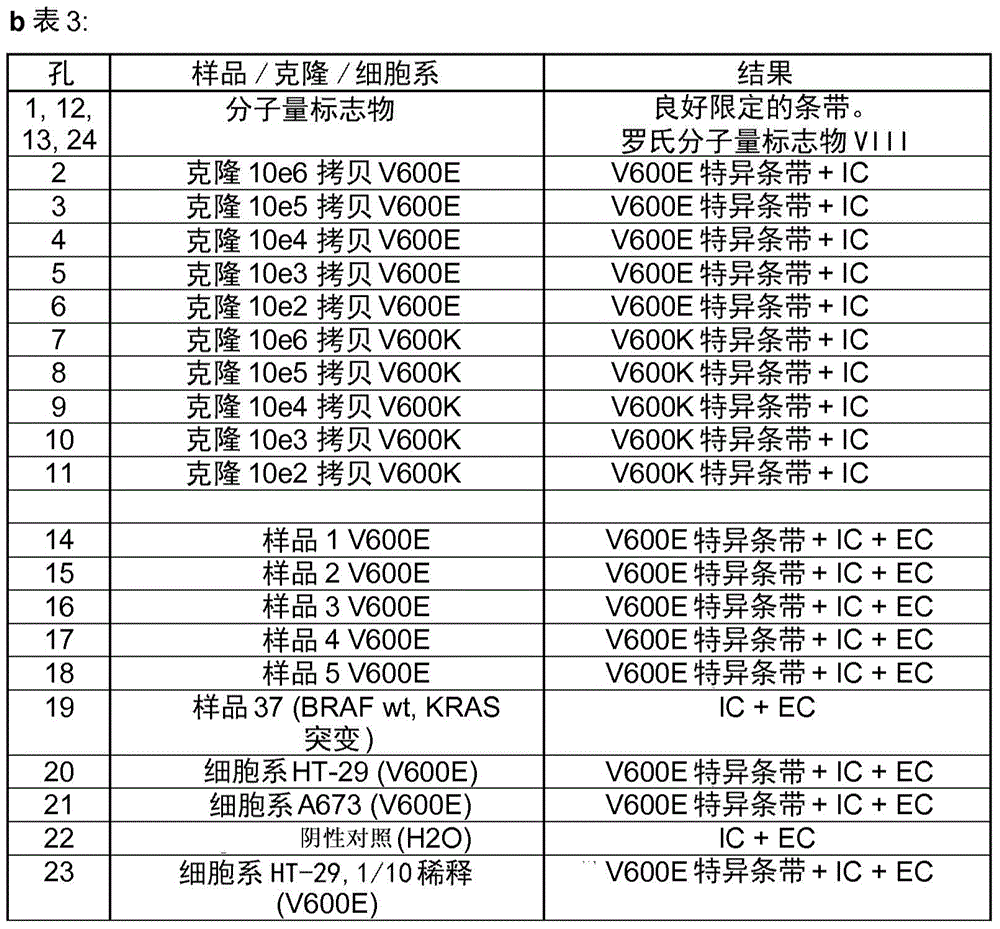
[0193] Will figure 2 The different ARMS products of the samples / clones or cell lines indicated in Table 3 were visualized in a 2% agarose gel in which the products of the different amplification reactions were analyzed.
[0194] The lengths of the different amplification products are as follows:
[0195] - Length of the mutation-specific bands corresponding to mutations V600E and V600K: 144bp;
[0196] -The length of the IC band: 101bp (amplified product o...
Embodiment 2
[0204] The compositions of amplification mixes 2 and 3 corresponding to PI3K are shown below. The remaining reaction conditions were the same as those shown in Example 1 for BRAF.
[0205]
[0206]
[0207] Corresponding to 5 μl of the cell line HTC 116 (0.2 ng / μl) containing the PI3K mutation H1047R after their respective amplifications with amplification mix 2 reagents, and subsequent hybridization of the products obtained to the probe microarray, and subsequent visualization (a), or 5 μl of clinical samples containing DNA at a concentration of 20 ng / μl (b) are shown in Figure 4 a) and 4b).
Embodiment 3
[0209] The contents of the amplification mix, forward and reverse primers necessary for internal control and extraction control amplification, and plasmids necessary for internal control amplification such as pBSK with insert, were obtained at the concentrations indicated below, unchanged the result of:
[0211] IC: internal control; EC: extraction control.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com