A method using SNPs to develop SNP-SSR molecular markers tightly linked to SNPs
A molecular labeling and labeling technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of no SSR marker positioning method, unfavorable gene fine positioning and cloning, and no physical location determination, etc., to shorten the research time Cycle, short cycle, effect of improving efficiency and quality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
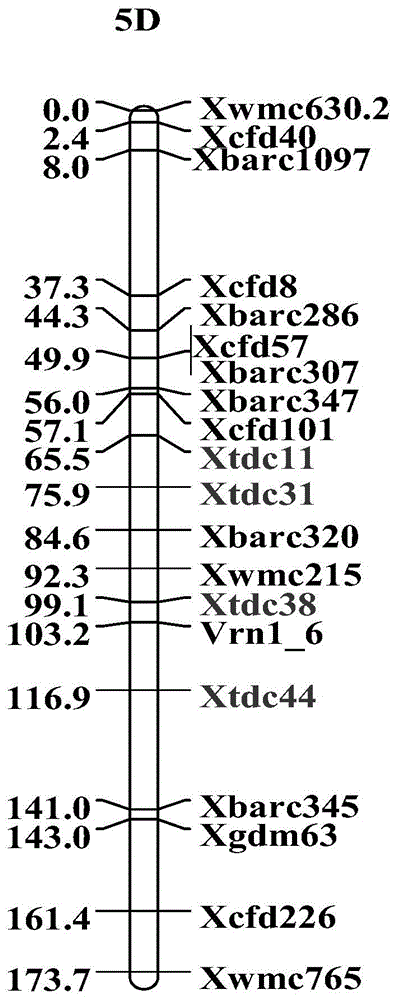
[0050] A method for obtaining a new SNP-SSR molecular marker closely linked to a SNP on wheat 5DL, comprising the following steps:
[0051] (1) According to the position of the molecular markers Xbarc320-Xwmc215 on the 5DL on the genetic map, it is determined that this segment is in the physical map of A. roughus http: / / probes.pw.usda.gov / WheatDMarker / The approximate position on the SNAP is AT5D4910-AT5D5010, which contains 90 SNP marker extension sequences;
[0052] (2) The extended sequences of the 90 SNP markers in step (1) were compared to the wheat D genome sequencing database (Jizeng Jia, Shancen zhao, Xiuying Kong, Yingrui Li, Guangyao Zhao, Weiming He, Rudi Appels. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation.Nature Letter,2013,496:91-95.) for local blast or http: / / wheat-urgi.versailles.inra.fr / Seq-Repository / BLAST Compare the provided wheat genome sequence database, and select sequence fragments with a length ranging from 5...
Embodiment 2
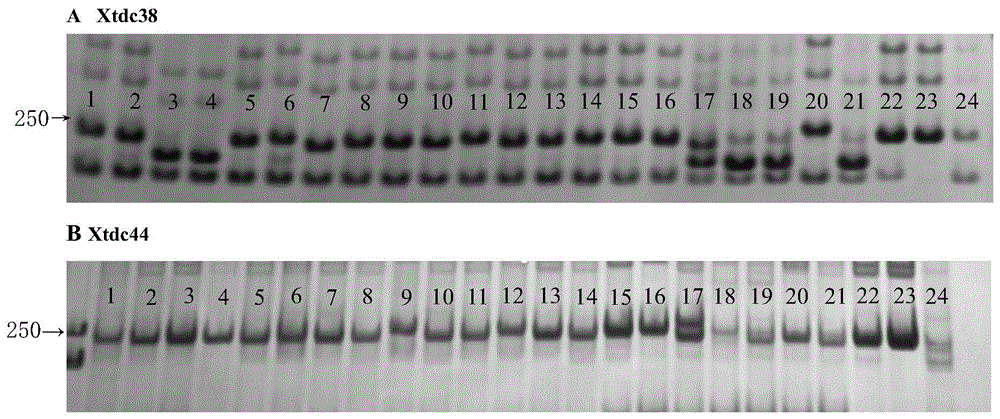
[0070] The application of the SNP-SSR molecular marker described in Example 1 in the detection of wheat varieties, the steps are as follows:
[0071] (1) Extract the genomic DNA of the wheat to be tested:
[0072] ① Take 3-4 young wheat leaves, put them in a centrifuge tube, put them in a container filled with liquid nitrogen, freeze them and grind them for 5-15 minutes;
[0073] ② Add 900 μL DNA extraction working solution preheated at 65°C, bathe in water at 65°C for 1 hour, shake gently 3 to 5 times during the water bath, and mix thoroughly;
[0074] ③After cooling at room temperature for 5 minutes, add an equal volume of phenol:chloroform:isoamyl alcohol (volume ratio 25:24:1) and mix well for 30 minutes, then centrifuge at 10000g for 20 minutes; take the supernatant and transfer it to a new centrifuge tube, and add an equal volume of chloroform: Isoamyl alcohol (volume ratio 24:1), after mixing gently, centrifuge at 10000g for 20min;
[0075] ④ Transfer the supernatant ...
Embodiment 3
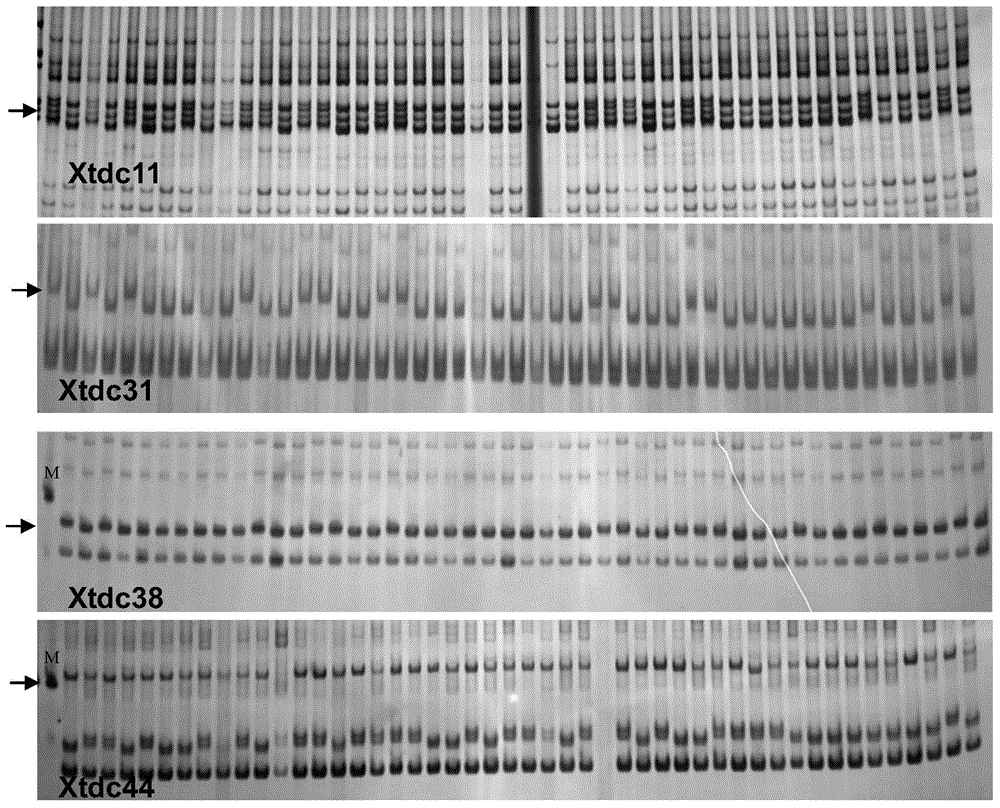
[0106] The application of the SNP-SSR molecular marker described in Example 1 in the construction of a molecular genetic map, using Yumai 57 as the male parent and Huapei 3 as the female parent to obtain F 1 , F 1 The haploid was induced by crossing wheat and maize, and the DH population containing 168 families was obtained through chromosome doubling; the application of new SNP-SSR markers closely linked to SNPs on wheat 5DL in the construction of molecular genetic maps was carried out, and the specific steps were as follows:
[0107] (ⅰ) Plant Yumai 57, Huapei 3 and the obtained DH populations in the field, and use the above-mentioned Triticarte Pty.Ltd (http: / / www.triticarte.com.au) on the leaves of the parents and 168 DH family plants The provided DNA extraction method extracts the DNA of each strain;
[0108] (ii) Perform PCR amplification with the detection primers of the new SNP-SSR molecular markers Xtdc11, Xtdc31, Xtdc38, and Xtdc44 respectively, and the reaction sys...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com