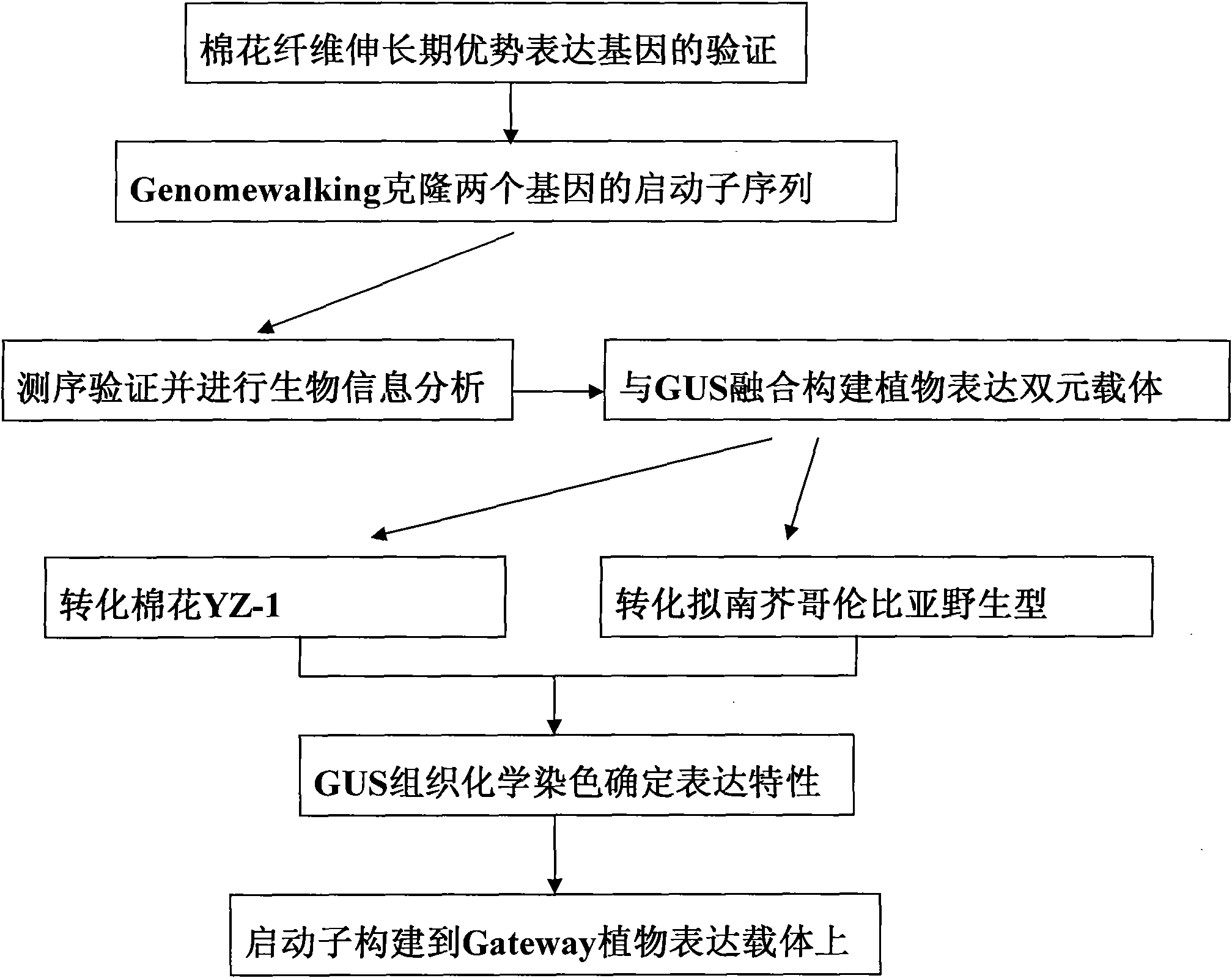
Two cotton fiber elongation stage preferential expression promoters and their application
A cotton fiber and promoter technology, applied in the field of plant genetic engineering, can solve problems such as lack of promoter resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Expression analysis of GbEXA1 and GbEXATR
[0025]The expression patterns of GbEXA1 and GbEXATR genes (gene accession numbers DQ912952 and DQ912951) were used to design primer pairs for 3'-UTR to verify their expression patterns by RT-PCR. The DNA sequence of the primer pair is as follows:
[0026] GbEXA1Sense: 5'CGATGGCAGGACTATCACAAAC3';
[0027] GbEXA1Anti: 5' TATAATATTGTCTTAAAAACTGGCC TCCTT 3'
[0028] GbEXATRSense: 5'GTGAAGAAAGGAGGCATCAG3';
[0029] GbEXATRAnti: 5'TCGAAAATACTTGCAAAAAT3'
[0030] The RT-PCR steps are as follows: extract the total RNA of different tissues: fibers of upland cotton TM-13, 5, 10, 15, 23DPA (days post anthesis) and ovules of 1-4 days before flowering; sea island cotton 3- Fibers of 5, 10, 15, 17, 23, 27 DPA of 79 and ovules 1-4 days before flowering; pollen, flowers, buds, leaves, ethylene-treated bell pedicle detachment layer, gibberellin-treated bell pedicle detachment layer , untreated bell stalk abscission layer, stem,...
Embodiment 2
[0032] Example 2: Obtaining the sequences of promoters PGbEXA1 and PGbEXATR
[0033] Primer GSP1 (gene special primer gene-specific primer) and primer GSP2 were designed with the full-length cDNA 3' end sequence and GenomeWalker TM Universal Kit (Protocol No.PT3042-2, purchased from Clontech Company, the U.S.) primer AP1, primer AP2 combined to carry out the cloning of the promoter (see the kit operation manual for specific steps), to obtain the primer pair shown below, its sequence is as follows Shown:
[0034] GbEXA1GSP1:CCACCGTAGAAGGTGGCATGGGCAGTTT
[0035] GbEXA1GSP2: ATTAGCACCAAGGAAAATGGAGTTGCAT
[0036] GbEXATRGSP1:CCACCGTAGAAGGTGGCATGGGCAGTTC
[0037] GbEXATRGSP2: ATTAGCACCAAGGAAAATGGAGTTGCAG
[0038] After gel electrophoresis, the target fragment was recovered with a gel recovery kit (the kit was purchased from Qiagene, Germany, Cat. No. 28704), and then TA cloned and sequenced. It is confirmed that the two promoters PGbEXA1 and PGbEXATR are 839bp and 1405bp in l...
Embodiment 3
[0039] Embodiment 3: Promoter PGbEXA1 and PGbEXATR drive the expression of GUS in cotton and Arabidopsis
[0040] Primers with restriction sites of Hind III and BamH I were designed to amplify the promoter, replace the 35S promoter driving GUS expression on the vector pBI121 (purchased from Clontech, U.S.), and transform the cotton line YZ-1 Check for fiber-specific promoters. The primer sequences are as follows (reverse primers for both promoters are the same):
[0041] PGbEXA1F: 5'agag aagctt AGTGCGAATAAAGAAGACCGCA'3
[0042] PGbEXATRF: 5'agag aagctt CACTTAAATTCTCAATAAAATTAGAAAAC'3
[0043] PEXR: 5'aga ggatcc TTGAGTAAAGAGCTAGCTAGCTCAAACAA'3
[0044] The enzyme cleavage sites in the above primers are indicated by lowercase English letters.
[0045] The method used for the cotton transgene involved in the present invention is the genetic transformation method mediated by Agrobacterium, and the Agrobacterium strain adopted is LBA4404 (Octopine Ti-plasmid deletion mutants of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com